x
Construct Design
The two constructs that we designed are detailed bellow. KDP is the potassium importer and GluR0 is the glutamate gated potassium channel. Details of the construction can be found here:
KDP
GluR0
KdpF-C Biobrick
Gene Selection
- Kdp is a well documented P-Type K+ ATPase found naturally in E.coli, used to actively pump ions into the cell.
- It consists of a 6-gene operon: F,A,B,C,D,E Where F-C are the functional membrane protein subunits, and D-E comprises a bacterial 2-component regulatory system.
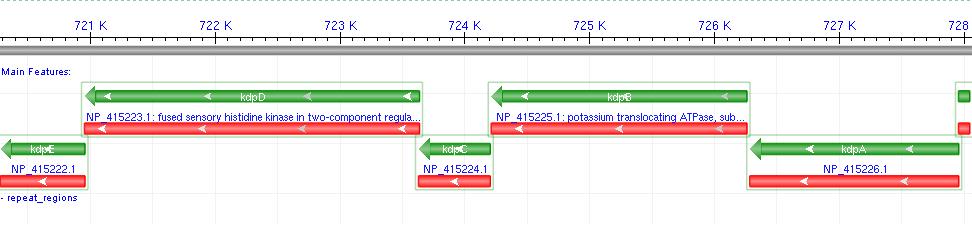
- Literature shows that Kdp acts as a high-affinity transport system, and works most effectively at low external potassium concentrations, where a change in ion flux would be most likely to produce a measurable voltage difference.
- The D-E 2-component system consists of a membrane protein turgidity sensor and a transcription factor. It controls Kdp operon expression in vivo, by reducing gene expression when turgor is high.
- Since we wish to over-express Kdp, we decided not to include the regulatory system in our biobrick. (Osmotic buffering would be used instead.)
Amplification from E.coli MG1655
- This was performed via PCR amplification of the genome template using the following primers:
-Forward: ATAT GAATTC ATAT TCTAGA TGAGTGCAGGCGTGATAACCGGCGTATT
EcoRI XbaI
-Reverse: CTCT CTGCAG CTCT ACTAGT TTATTCATCAAGTTTATCCAGCGCCAGAT
PstI SpeI
- Primer overhangs incorporated the biobrick prefix and suffix into the section, restriction sites shown in bold .
- The result of this PCR is shown below:
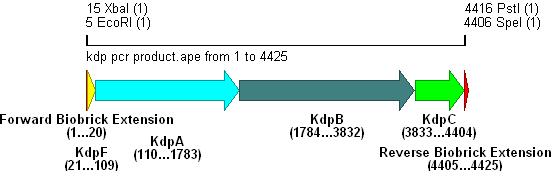
Integration into Vector
- The vector used was low copy-number plasmid [http://partsregistry.org/Part:pSB4C5 pSB4C5], with chloramphenicol resistance and a death gene as selection markers.
- Kdp PCR product and pSB4C5 were both cut with EcoRI & SpeI, (vector backbone was dephosphorylated to prevent circularisation) then ligation into the vector can occur as shown.
- . . . . . . . . . . . . . . . . . . . . . . . .
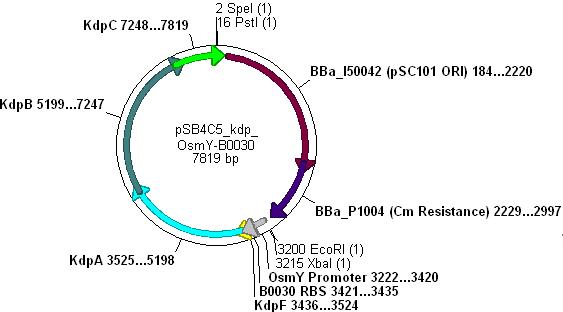
- Note: pSB4C5_Kdp biobrick plasmid has no promoter/RBS and so Kdp is not expressed in transformants.
Promoter+RBS Biobrick
Promoter and RBS Selection
Promoter
- The promoter chosen for use with Kdp was OsmY [http://partsregistry.org/Part:BBa_J45992 (Part BBa_J45992).]
- It is a stationary phase promoter, and since we require high cell densities in our final "voltage measurement" medium, we want Kdp to only be expressed in stationary phase.
- This will reduce the metabolic and osmotic stress on dividing cells in exponential phase.
Ribosome Binding Site
- Three different strength RBSs were investigated, [http://partsregistry.org/Part:BBa_B0030 B0030], [http://partsregistry.org/Part:BBa_B0031 B0031] and [http://partsregistry.org/Part:BBa_B0032 B0032].
- B0030 is the strongest(15bp length), B0031 medium(14bp) and B0032 weakest(13bp).
- Investigating three will help us determine the optimum levels of Kdp expression.
Amplification from E.coli MG1655
- These parts were extracted using PCR from the Registry of Standard Biological Parts. However, the RBS biobricks are so small that we built their sequences into the reverse primers.
- The primer sequences used are:
-Forward: CTAT GAATTC ATAT TCTAGA GCTGGCACAGGAACGTTATCC (All OsmY-RBS constructs)
EcoRI XbaI
-B0030 Reverse: CGCG CTGCAG CTCT ACTAGT (TTTCTCCTCTTTAAT)TTGTTAAATATAGA
PstI SpeI B0030
-B0031 Reverse: CTCT CTGCAG CTCT ACTAGT (GGTTTCCTGTGTGA)TTGTTAAATATAGAT
PstI SpeI B0031
-B0032 Reverse: CTCT CTGCAG CTCT ACTAGT (CTTTCCTGTGTGA)TTGTTAAATATAGATCA
PstI SpeI B0032
- PCR with these primers creates three different promoter-RBS biobrick parts (OsmY-B003x):
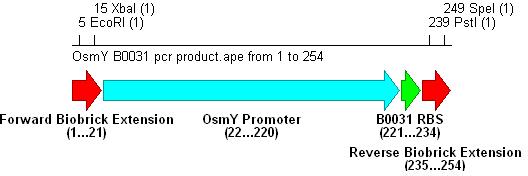
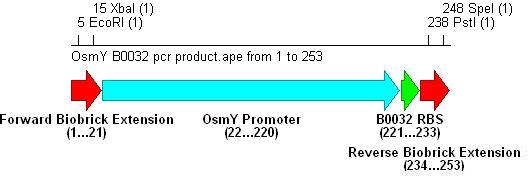
Integration into Vector
- The vector used was low copy-number plasmid [http://partsregistry.org/Part:pSB4C5 pSB4C5], with chloramphenicol resistance and a death gene as selection markers.
- OsmY-B003x and pSB4C5 were both cut with XbaI & SpeI, (vector backbone was dephosphorylated to prevent circularisation) then ligation into the vector can occur as shown:
- . . . . . . . . . . . . . . . . . . . . . .
- Note: This promoter-RBS construct did not cause unwanted transcript problems because there are many double terminators scattered throughout the pSB4C5 backbone.
Combination of Kdp, OsmY and B003x
- This will create a functional biobrick plasmid in which Kdp is overexpressed only in stationary phase of growing cells.
- Cut pSB4C5-Kdp with XbaI and PstI
- Cut pSB4C5-OsmY-B003x with PstI first, then SpeI, in order to make sure Pst cuts correctly.
- Ligation will form the following functional plasmid:
GluR0 Biobrick
Gene Selection
- In order to create a measurable voltage change when a chemical was "recognised" we decided to use an ionotropic ligand-gated potassium efflux channel that binds glutamate. This also simulates the action of glutamate as a neurotransmitter in the CNS.
- The gene chosen comes from the cyanobacteria Synechocystis sp. PCC 6803. This species is gram negative (similar to E.coli) and is used as a paradigm for evolutionary research concerning the AMPA receptor proteins.
- GluR0 is a well characterised glutamate-gated K+ membrane channel, which has a considerable degree of structural and functional homology to rat neurone GluR2 AMPA receptors, see the paper [http://www.nature.com/nature/journal/v402/n6763/full/402817a0.html Functional Characterisation of a Glutamate-gated Potassium Channel]
DNA Synthesis
- The protein sequence was obtained via NCBI from the [http://www.ncbi.nlm.nih.gov/entrez/viewer.fcgi?tool=portal&db=protein&term=&query%5Fkey=2&dopt=gp&dispmax=20&page=1&qty=1&WebEnv=0Z1aMb1oSyT%5FAsbY%5Fe8SHH5%5FIGnme2UYVwupBbQ8pXgSmignz7xFMxGxDxnS%5FdHJl5fpNJ1RyQVTXT3f%404A8B168688057500%5F0127SID&WebEnvRq=1 Synechocystis sp. PCC 6803 genome].
- The gene sequence could not be directly used because of codon optimisation problems (Synechocystis uses many codons that are "rare" in E.coli"
- The protein sequence was back-translated to DNA using GeneDesigner™ and codons were assigned using the E.coli usage table.
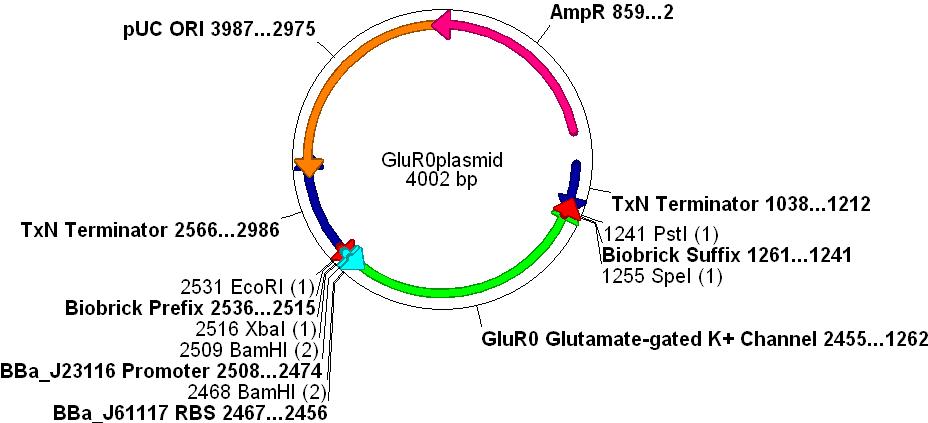
- The promoter [http://partsregistry.org/Part:BBa_J23116 BBa_J23116], RBS [http://partsregistry.org/Part:BBa_J61117 BBa_J61117] and Biobrick prefix and suffix sequences were added to the design.
- Finally, unwanted restriction sites within the gene(EcoRI, XbaI, SpeI and PstI) were manually removed by selecting alternative codons for any given amino acid.
- The gene was synthesised and sequenced by DNA2.0 into one of their standard vectors.
x