|
THIS PAGE IS A DRAFT, COPY OF THE OFFICIAL PAGE, EXISTING ONLY IN RDER TO BE ANNOTATED FOR CORRECTIONS AND IMPROVEMENT OF THE OFFICIAL PAGE
Introduction
- Aims of the modeling part
- First approach proposed
- Hill functions
- first model + score function
- bibliography
- findparam
- experiments
- Second approach
- bibliography
- equations
- results
- experiments
- Continue the previous model
- Synchronyzation
- Estimation of the FIFO processes
- Stochastic modeling (Gilespie)
- Test of robustness
- Enhancing the system
- Better FIFO behaviour
- Other interactions to increase the robustness
Roadmap
If you want to have a look at our modeling notebook: Notebook
An Oscillatory Biological Model
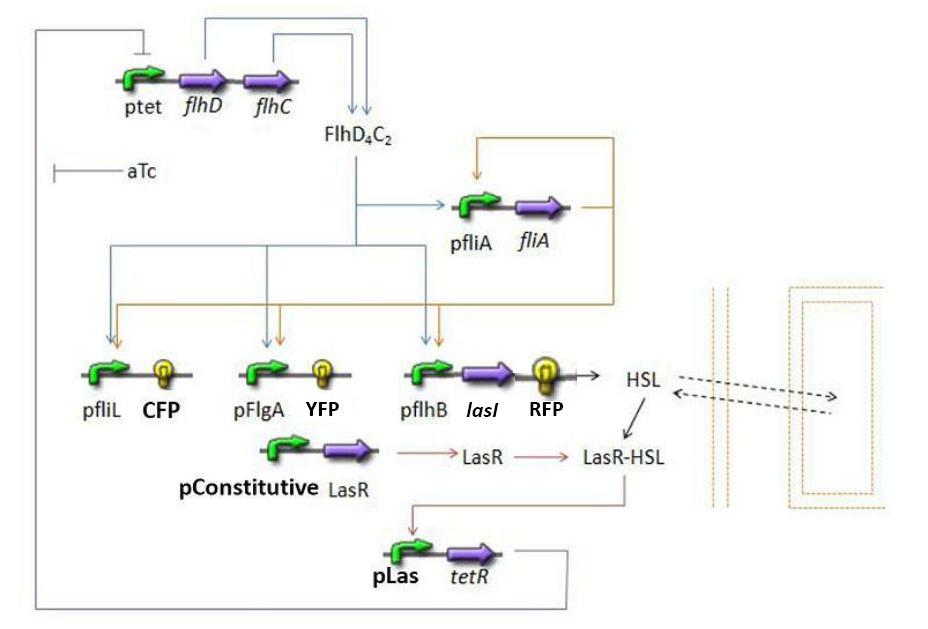 Overall Model Overall Model Introduction
The goal here is to present the differential equations we used in the system modelization. At each step, we shall describe why we chose this precise model, its drawbacks and possible improvments, the parameters involved and enventually a biologically coherent value.
The key problem with a dynamic system consists in the fact that adding a new equation gives a more detailed idea of the overall process, but one looses precision by doing so since new parameters appear.
In that respect (in all cases but one) we chose not to model the mRNA steps, that is translation and transcription. We then assumed that we could act as if a protein would directly act upon the follower, without loosing too much precision.
Is is worth to comment somewhere more on this assumption. Why do you assume it will not change much the dynamics?''
This first approach refers to a set of two cells. We shall examine later on what happens if we put more cells together.
It seems important to understand that with this approach, we did not intend to design the most accurate model possible. We found essential to chose only parameters that we could determine and command in the wet lab.
Please introduce already in the introduction what will be your two models, in a few words the differences
Bibliography
Much of our inspiration comes from four articles to which we shall refer in the next subsections :
- [1] Shiraz Kalir, Uri Alon. Using quantitative blueprint to reprogram the dynamics of the flagella network. Cell, June 11, 2004, Vol.117, 713-720.
- [2]Jordi Garcia-Ojalvo, Michael B. Elowitz, Steven H. Strogratz. Modeling a synthetic multicellular clock : repressilators coupled by quorum sensing. PNAS, July 27, 204, Vol. 101, no. 30.
- [3]Nitzan Rosenfeld, Uri Alon. Response delays and the structure of transcription networks. JMB, 2003, 329, 645-654.
- [4]Nitzan Rosenfeld, Michael B. Elowitz, Uri Alon. Negative autoregulation speeds the response times of transcription networks. JMB, 2003, 323, 785-793.
- [5]S.Kalir, J. McClure, K. Pabbaraju, C. Southward, M. Ronen, S. Leibler, M. G. Surette, U. Alon. Ordering genes in a flagella pathway by analysis of expression kinetics from living bacteria. Science, June 2001, Vol 292.
Equations
- Steps
- flhDC → fliA (1)
- flhDC → fliL → Fluorescent Protein 1 (FP1) (2)
- flhDC → flgA → Fluorescent Protein 2 (FP2) (3)
- flhDC → flhB → Fluorescent Protein 3 (FP3) (4)
- flhDC → flhB → lasI (5)
- fliA → fliL → Fluorescent Protein 1 (FP1) (6)
- fliA → flgA → Fluorescent Protein 2 (FP2) (7)
- fliA → flhB → Fluorescent Protein 3 (FP3) (8)
- fliA → flhB → lasI (9)
Those are not properly the equations, but a description as chemical reactions that you will then translate into equations by doing proper assumptions. Those are, indeed, the "steps" that are represented graphically above. I would suggest that for clarity you first list here ALL the steps reactions (numbering them) and then only proceed with the modeling of each group of steps as you do below.
The transcription hierarchy of the flagella genes have been studied in [1]. It resulted that the promoter activity of the seven class 2 operons, among which fLiL, flgA, flhB, may be mathematically described in that way :
What is a class 2 operon ?
This is a linear equations, a comment would be justified. It is not a trivial issue.
where [X] denotes the effective protein-level activity.
For each operon, Shiraz Kalir and Uri Alon came up with numerical values of β and β', available in [1].
Furthermore, the protein-level activity can be presented (for a more detailed presentation, see [4]) as
Thus :
Discussion and Assumptions
- As presented in the first equation below, there is a retroaction from fliA over fliA. In a first approach, we decided not to take into account this interaction, that is setting β'FliA to zero. In a second approach, we will consider this retroaction, and find a way to describe it mathematically.
- Another step consists in choosing the proper units, so as to have a homogenous set of parameters.
</br> Again, it is not at all a trivial fact that the equations here are linear. Discuss.
- Steps
- lasI → HSLext (10)
- lasI → HSLint (11)
- Discussion and Assumptions
- In a first approach, we have assumed that HSL could be modelized in the same fashion as AHL. The process was well detailed in [2] and we adapted their model to our present situation. Furthermore, a biologically coherent set of parameters is also given in this article. We decided then to use them as well for our simulation.
It is good to give the reference, but for the readability of your presentation you should include here also briefly the justification of your model. Too rapid just to refer to the bibliography.
- Steps
- HSLint → tetR mRNA (12)
- tetR mRNA → tet R (13)
- Discussion and Assumptions
- Here is the only case where we have not yet found out how we could manage to skip the mRNA step. However, this is acceptable since every parameter should be available. Then, (12) represents the binding and transcription steps ; (13) represents the translation step.
Could you discuss why? It is not clear to me straight on.
- The effect of HSL as well as the link between the mRNA activity and protein activity is well studied in [2]. We were the able to adapt the introduced situation to our system.
- The ρ parameter corresponds to maximal contribution to tetR transcription in presence of saturating amounts of HSL. Yet, to begin with, we have used the parameter given in the text. We shall then find a more accurate parameter corresponding to our system. The HSL activation is hereby chosen to follow Michalelis Menten kinetics since we assumed that there would be no cooperativity.
- Finally, we assumed that lasR would exist permanently in the medium in sufficient amounts, so that we needn't take it into account mathematically speaking.
- Discussion and Assumptions
- To determine this part of the model, we used [4] where the influence of tetR is described.
- Finally, to close the loop, we saw the necessity to use a little refinment. the influence of tetR over pTet (pTet being the promoter of flhDC) is a repression. To express the fact that a certain concentration is needed for the expression of flhDC, we used a step function. This choice was influenced by [3]. To fully define this function, two parameters are needed, so as to describe the threshold when the step has to occur, and the initial value of this step. We are facing two solutions. The more accurate procedure would consist in getting those values from the wet-lab. However, we may also look it up in the literature. Anyhow, as presented in the conclusive part, there is a really wide range of parameters that enable the system to oscillate.
What is the function you are using for F()?
- The key problem was that those two parameters were to be determined by the overall evolution of tetR, and the program cannot wait for tetR to be entirely determined to evaluate flhDC, since we have a loop. We then decided that these parameters could be obtanied by lab experiments.
Parameters summary
- Awaiting for experimental values, since it seemed coherent by reading articles, we have chosen to set every degradation rate to 1.
| Parameter Table
|
| Parameter
| Parameter in code (cell i)
| Meaning
| Value
| Unit
| Source
| Accuracy
| Comments
|
| γFliA
| g[1 + 8 x (i-1)]
| Degradation rate
| 1
|
| [1]
|
|
|
| βFliA
| B(1,i)
| FlhDC activation coefficient
| 50
|
| [1]
|
|
|
| β'FliA
| b(1,i)
| FliA activation coefficient
| 0
|
| [1]
|
|
|
| γFP1
| g[2 + 8 x (i-1)]
| Degradation rate
| 1
|
| [1]
|
|
|
| βFP1
| B(2,i)
| FlhDC activation coefficient
| 1200
|
| [1]
|
|
|
| β'FP1
| b(2,i)
| FliA activation coefficient
| 250
|
| [1]
|
|
|
| γFP2
| g[3 + 8 x (i-1)]
| Degradation rate
| 1
|
| [1]
|
|
|
| βFP2
| B(3,i)
| FlhDC activation coefficient
| 150
|
| [1]
|
|
|
| β'FP2
| b(3,i)
| FliA activation coefficient
| 300
|
| [1]
|
|
|
| γFP3
| g[4 + 8 x (i-1)]
| Degradation rate
| 1
|
| [1]
|
|
|
| βFP3
| B(4,i)
| FlhDC activation coefficient
| 100
|
| [1]
|
|
|
| β'FP3
| b(4,i)
| FliA activation coefficient
| 350
|
| [1]
|
|
|
| γlasI
| g[4 + 8 x (i-1)]
| Degradation rate
| 1
|
| [1]
|
|
|
| βlasI
| B(4,i)
| FlhDC activation coefficient
| 100
|
| [1]
|
|
|
| β'lasI
| b(4,i)
| FliA activation coefficient
| 350
|
| [1]
|
|
|
γHSLint
| g[5 + 8 x (i-1)]
| Degradation rate
| 1
|
| [1]
|
|
|
βHSLint
| B(5,i)
| Production rate
| 0,01
|
| [2]
|
|
|
| δint
| D(1,i)
| Diffusion rate in the cell
| 2
|
| [2]
|
|
|
γHSLext
| g[6 + 8 x (i-1)]
| Degradation rate
| 1
|
| [1]
|
|
|
| δext
| D(2,i)
| Diffusion rate out off the cell
| 2
|
| [2]
|
|
|
| γtetR mRNA
| g[6 + 8 x (i-1)]
| Degradation rate
| 1
|
| [1]
|
|
|
| ρ
| R(i)
| Maximal contribution to tetR in the presence of saturating amounts of HSL
| 20
|
| [2]
|
|
|
| θ
| 0(i)
| Ratio between mRNA and protein lifetimes
| 1
|
| [2]
|
|
|
| γflhDC
| g[8 + 8 x (i-1)]
| Degradation rate
| 1
|
| [1]
|
|
|
| τ
| T(i)
| Threshold for tetR repression
| (20000)
|
| lab
|
|
|
| ∆
| Pa(i)
| Promoter activity without the repressor
| (100)
|
| lab
|
|
|
Graph screenshots
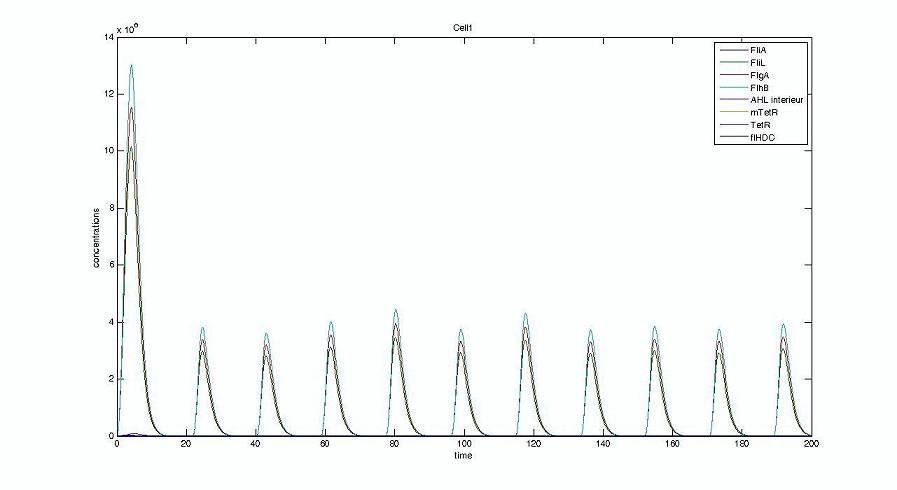 Cell1 in a 2 cells model : oscillations and a glimpse of FIFO Cell1 in a 2 cells model : oscillations and a glimpse of FIFO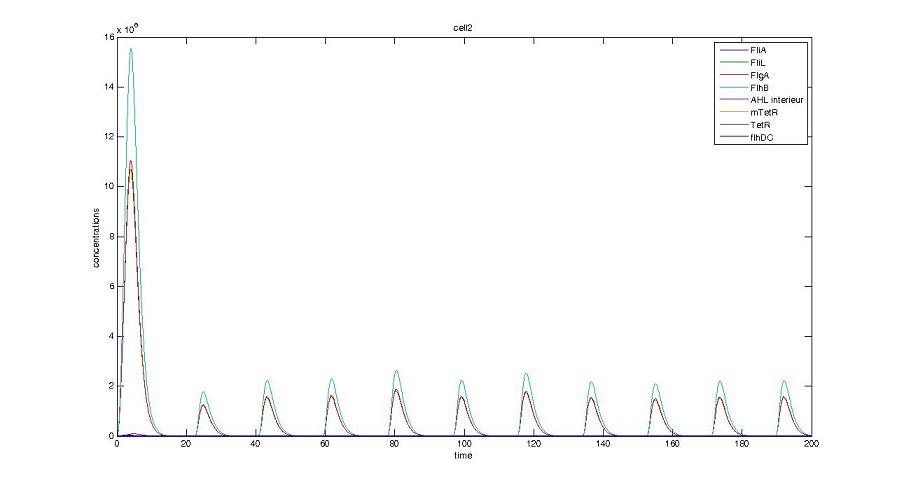 Cell2 (noise added to set the parameters) in a 2 cells model : oscillations and a glimpse of FIFO Cell2 (noise added to set the parameters) in a 2 cells model : oscillations and a glimpse of FIFO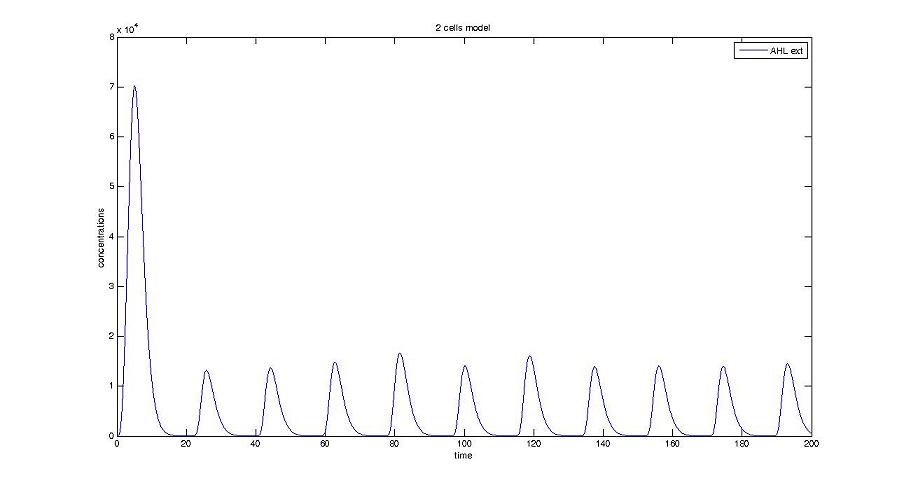 HSL ext in a 2 cells model : oscillations HSL ext in a 2 cells model : oscillations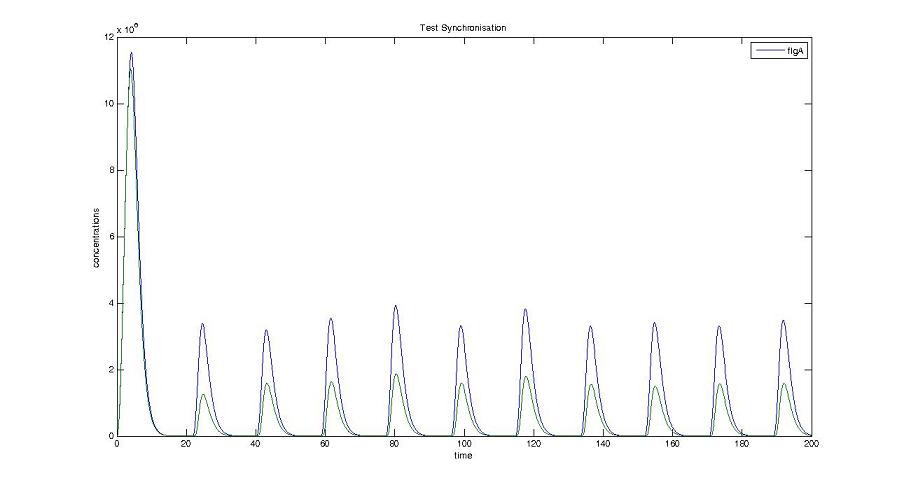 Synchronisation : flgA in both cells Synchronisation : flgA in both cells Synchronisation : fliL in both cells Synchronisation : fliL in both cells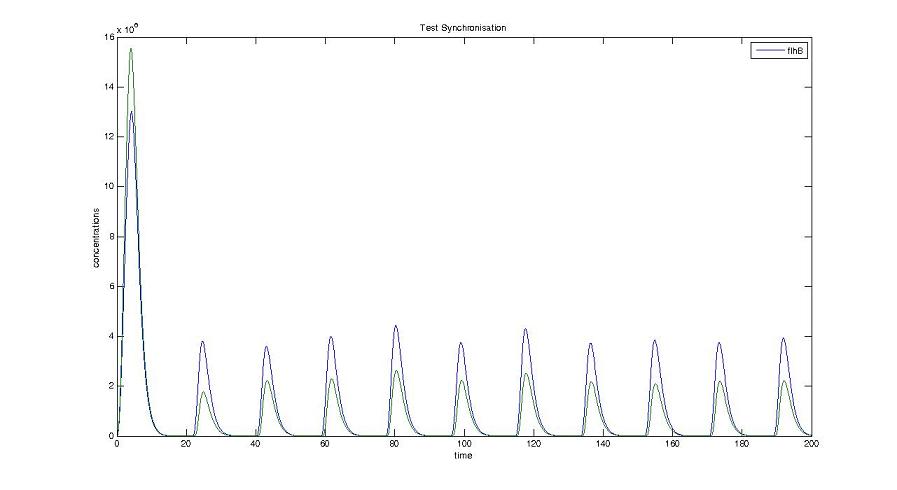 Synchronisation : flhB in both cells Synchronisation : flhB in both cells Corresponding codes
- To change the parameters, you have to use this file, and refer to the previous array : Model
- To launch the numerical simulation and plot the curves : Plot the curves
- To compute the numerical simulations (does not provide any visual output, but necessary for the previous script) : Numerical simulations
- To add noise to deterministic parameters : Noise function
- To define all the parameters in a row : Parameters
Discussion
- In [1], we had trouble deciding whether the parameters we used were coherent or not, and we spent some time checking abouts units. Thus, we had to decide how we shall set our time unit.
- We found a seemingly acceptable answer using elements from [1] and [5].
Good section for discussing relationship between model quantities and whole population measurements. You should explain better the purpose of this part however. What are the conditions of measurement. Why the optical density. What is $GFP_{1cell}$. Protein level activity is the concentrationof the protein? What is $[Y]$. Describe more how to get the final relationship between $T_{equation}$ and $T_{real}$.
- If you compare these results with our program codes, you might see that we did not take the α factor into account. However, we assumed that we could use it so as to produce our time scale. In fact, we are given the time taken by a population to double its size and since, we know that
| From equations to reality
|
| Equations
| Reality
|
| Tdouble-eq = ln2 / α
| Tdouble
|
- This enabled us to set a real x-axis unit (we had decided for practical reasons to set α to 1).
- Finally, we have compared our results with the experiments carried out in [5]. It appears that the timescales to turn the genes 'ON' are similar !
Explain the comparison. It is not clear where are the time scales you compared.
Concerning the parameters taken from [2], it has to be noticed that output units are arbitrary. Nonetheless, as the degradation rate is also 1, as in [1], one can infer that the transformations (to obtain the 'real' time and Fluorescence units) are the same in [2] than in [1].
Thanks to that assumptions, we are finally able to end up with a quantitative description of the biological system, instead of just a qualitative one.
First Mathematical Approach
Introduction
This description of the approaches should come earlier. It is not clear reading you if you are speaking of the model presented above, or of something else you did before. I think this part that presents your work on modeling should come before the detailed developments of above. In the introduction to help the reader understand where you are heading.
As a first approach, we had decided to take into account the binding to the promoters steps. Moreover, the transcription rates were expected to be Hill functions. Obvisouly, this modeling requires a huge number of parameters. To obtain them, we had planed to devise specific experiments (described below).
Nonetheless, after reading some more articles, we have decided to change several assumptions of the modeling choice. Therefore, we have devised a perhaps more biologically relevant framework (see above).
This part describes in detail the first approach and the codes that have been produced.
First Approach
As a first approximation, we have proposed a set of 5 ordinary differential equations, without taking into account the transcription step. Besides, we have had not introduced yet a synchronizaton module. Therefore, the repression of FlhDC is directly modeled by the presence of the 'Z3' gene (that is the last that is turned on).
In this framework, we have found parameters that have provided oscillations as well as a function that automatically detects whether the output of the ode system is oscillating. This has allowed to screen a little the parameters used, in order to evaluate the robustness of the system.
The methods employed are described there : First Approach.
More precise Bio-Mathematical Description
After trying to obtain oscillations from a simple model, we have tried to described more precisely the studied system. Therefore, we have obtained the following formalism : Bio-Mathematical Description.
Bibliography
In order to choose a proper modeling approach for our system, we have decided to list all the chemical reactions we will take into account. Afterwards, we will find the needed parameters reading articles or devising the required experiments.
An overview of the work that has to be done can be found here : Parameters we have to use.
Estimation of parameters
Then, we will need many parameters to fully desribe the system according to the asumptions of the previous section. A natural way to have access to their value, after searching in the litterature, is to devise specific experiments. As a consequence of the characterization of the promoters activity, some Hill functions could be obtained.
Thus, we have described the experimental approach required : Estimation of the parameters.
Nonetheless, as mentioned above, we have changed the way to model the biological reactions. As a result have stopped investigating in this way to focus on the An Oscillatory Biological Model.
|