Team:Heidelberg/Parts/Characterization
From 2008.igem.org


Part Characterization
Bacteriophage lambda cI ([http://partsregistry.org/wiki/index.php?title=Part:BBa_K150004 BBa_K150004])
cI protein - an introduction
Bacteriophage λ cI exists as an inactive monomer at very low concentrations (<10-9 M) but forms functional homodimers at physiological concentrations that remarkably lack a global symmetry [1]. It consists of 236 amino acids, although 237 amino acids are translated since the initiator methionine is removed by the host. Although λ cI is commonly called a repressor because of its negative regulatory functions at oL and oR, cI protein acts also as a positive regulator of gene transcription and can activate transcription of its own gene through the pRM promoter in bacteriophage λ. In genetic and biochemical studies as well as through the crystal structure it has been shown, that the carboxy-terminal domain of the cI protein contains the major sites for dimerization and oligomerization [2, 3]. The cI protein has a two-domain structure with an N-terminal portion involved in DNA binding, and a C-terminal domain that mediates dimer formation, dimer-dimer interaction, and self-cleavage. The self-cleavage reaction is triggert when the lysogenic cell suffers DNA damage and depends upon an activated form of the bacterial RecA protein [2, 4-6]. The ‘hinge region’ connecting both domains contains a conserved site that can undergo this RecA-mediated autodigestion resulting in inactivation of the repressor by separating the N-terminal from the C-terminal domain of the repressor [4, 7]. The cI protein binds symmetrically to DNA, so that each amino-terminal domain contacts a similar set of bases. The N-terminal DNA-binding domain is made up of 5 α-helices of which helix 2 and helix 3 (the helix-turn-helix motif) are involved in nucleotide sequence specific DNA recognition and binding to the major groove of DNA [8-11]. [back]
Function of cI protein in bacteriophage λ
The cI repressor in bacterophage λ is the key component of a ‘genetic switch’ that enables the phage to transition from lysogenic growth to lytic development. The cI protein binds the oL and oR operator which overlap with the pL and pR promoter (the lytic promoters). This binding allows the maintenance of the lysogenic state to be governed by cI alone. As soon as cI is inactivated, e.g., by the SOS response after UV damage or other agents that cause DNA damage, the lytic development follows [12]. The two operators oL and oR both contain three binding sites for cI protein. In each case, site 1 (i.e. oL1 and oR1) has a ~10-fold greater affinity than the other sites for cI protein. The repressor, therefore, always binds to oL1 and oR1 first and than to the other sites in the operator in a cooperative manner [13, 14]. The carboxy-terminal domains of the repressor dimers mediate this cooperativity which improves the specificity and strength of the cI DNA binding, enabling strong repression of the lytic promoters [15]. Furthermore, another cooperative interaction between these two sets of tetramers bound to oL and oR, 2.4 kb apart, leads to the formation of a DNA loop held by a cI octamer, i.e. two interacting tetramers, that enhances repression of the early promoters [16-19]. In this DNA-multiprotein complex, the cI dimer bound at oR2 represses pR and at the same time also stimulates pRM transcription, thus activating cI synthesis in a repressed prophage by a positive autoregulatory loop [20-22]. As the cI concentration increases because of pRM activation, two additional cI dimers are recruited to bind oL3 and oR3 to further stabilize the oL – oR loop. In this context, cI overexpression is prevented by the binding of cI to oR3 which represses pRM [17, 19]. This positive and negative autoregulation at pRM by cI ensures a narrow range of cI repressor level to be maintained, which is optimum for stable lysogeny but is at the same time adjusted low enough for efficient induction of the lysogen. By repressing transcription from the pR promoter not only expression of genes in that operon is inhibited but also phage DNA replication by preventing transcriptional activation of λ ori, the site where phage DNA replication is initiated [23]. Even if O and P functions are present this inhibition occurs and appears to be critical for establishing a lysogen [24, 25]. The stable lysogen produces sufficient repressor not only to block prophage lytic development but also to block lytic development of any extraneous infection phage, thus imparting immunity to the lysogen against lytic superinfection [26]. [back]
Characterization of cI ([http://partsregistry.org/wiki/index.php?title=Part:BBa_K150004 BBa_K150004])
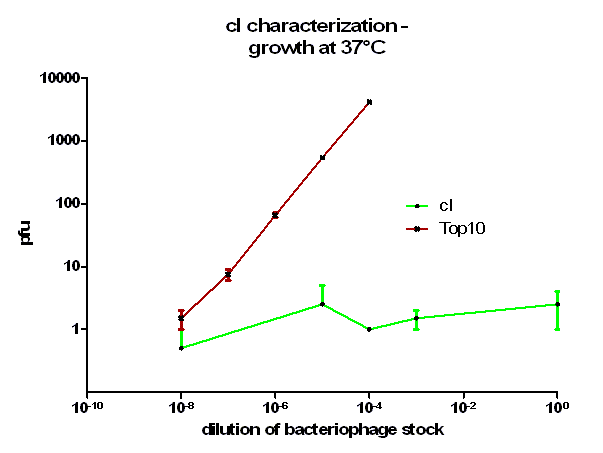
For the characterization of λ cI protein its ability to repress lytic development of bacteriophage λ was measured over a range of temperature from 28 °C to 42 °C in a phage burst experiment. If the used cI is functional no plaques should appear in a lawn of cells harboring this cI when infected with a phage that lacks its natural cI. In control cells in contrast every of this virulent particles should lead to a plaque.
Protocol
Preparation of plating bacteria
A overnight culture of the appropriate E. coli strain was grown in LB medium containing 10 mM MgSO4 and 0.2 % maltose at 30 °C to reduce the amount of cell debris in the medium. For this characterization E. coli TOP 10 and E. coli TOP 10 with transformed cI were used. The added maltose leads to a substantial induction of the maltose operon including the lamb gene, which encodes the cell surface receptor to which bacteriophage λ binds. After harvesting the cells at 3000g for 10 minutes they were resuspended in 10 mM MgSO4 and diluted to a final concentration of 2.0 OD600. The suspension of plating bacteria was stored at 4 °C for up to 1 week.
Bacteriophage λ plaque assay
Tenfold serial dilutions of the bacteriophage λ stocks were prepared. In this case a bacteriophage mutant was used lacking a functional cI which would therefore always lead to plaques. From these dilutions 100 µl were mixed with the same amount of plating bacteria, incubated for 20 minutes at 37 °C to allow the bacteriophage particles to adsorb to the bacteria, this mixture than added to 3 ml molten top agar which was kept liquid at 48 °C and the entire contend poured onto a agar plate. After harden of the top agar the inverted plates were incubated at indicated temperatures over night. On the next day plaques could be counted.
Results
As it can be seen in figure 3 – 5 the cells harboring the part [http://partsregistry.org/wiki/index.php?title=Part:BBa_K150004 BBa_K150004] are not lysed by the used phage. This means that the constructed cI protein generator successfully expresses high levels of cI protein which is functional. Through the use of different temperatures it could be shown that the ability of the used cI protein to repress lytic development is not dependent of the temperature.
[back]
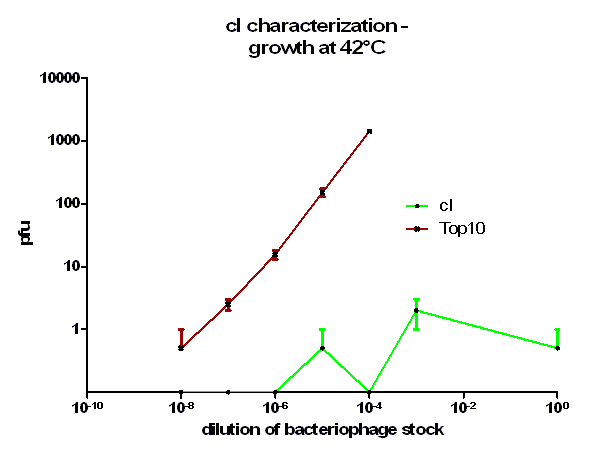
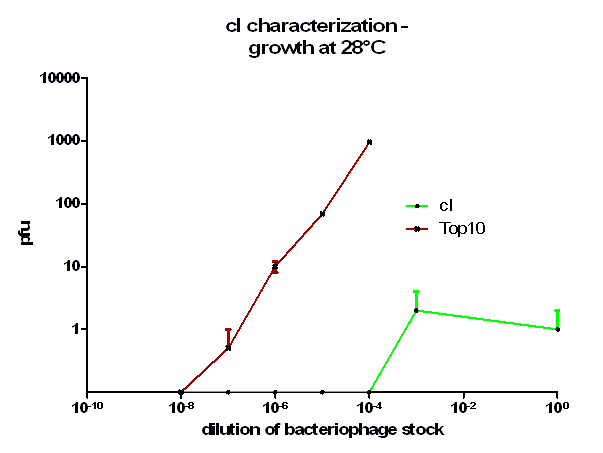
[back]
References
[1] S. Stayrook, P. Jaru-Ampornpan, J. Ni, A. Hochschild, M. Lewis, Crystal structure of the lambda repressor and a model for pairwise cooperative operator binding, Nature 452 (2008) 1022-1025.
[2] C.O. Pabo, R.T. Sauer, J.M. Sturtevant, M. Ptashne, The lambda repressor contains two domains, Proc Natl Acad Sci U S A 76 (1979) 1608-1612.
[3] C.E. Bell, P. Frescura, A. Hochschild, M. Lewis, Crystal structure of the lambda repressor C-terminal domain provides a model for cooperative operator binding, Cell 101 (2000) 801-811.
[4] J.W. Little, Autodigestion of lexA and phage lambda repressors, Proc Natl Acad Sci U S A 81 (1984) 1375-1379.
[5] R.T. Sauer, M.J. Ross, M. Ptashne, Cleavage of the lambda and P22 repressors by recA protein, J Biol Chem 257 (1982) 4458-4462.
[6] R.T. Sauer, C.O. Pabo, B.J. Meyer, M. Ptashne, K.C. Backman, Regulatory functions of the lambda repressor reside in the amino-terminal domain, Nature 279 (1979) 396-400.
[7] J.W. Little, LexA cleavage and other self-processing reactions, J Bacteriol 175 (1993) 4943-4950.
[8] C.O. Pabo, M. Lewis, The operator-binding domain of lambda repressor: structure and DNA recognition, Nature 298 (1982) 443-447.
[9] L.J. Beamer, C.O. Pabo, Refined 1.8 A crystal structure of the lambda repressor-operator complex, J Mol Biol 227 (1992) 177-196.
[10] A. Hochschild, Transcriptional activation. How lambda repressor talks to RNA polymerase, Curr Biol 4 (1994) 440-442.
[11] C.O. Pabo, R.T. Sauer, Transcription factors: structural families and principles of DNA recognition, Annu Rev Biochem 61 (1992) 1053-1095.
[12] A. Hochschild, The lambda switch: cI closes the gap in autoregulation, Curr Biol 12 (2002) R87-89.
[13] A.D. Johnson, B.J. Meyer, M. Ptashne, Interactions between DNA-bound repressors govern regulation by the lambda phage repressor, Proc Natl Acad Sci U S A 76 (1979) 5061-5065.
[14] M. Ptashne, Genetic switch: phage lambda revisited, 2nd ed., Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, 2004.
[15] I.B. Dodd, K.E. Shearwin, J.B. Egan, Revisited gene regulation in bacteriophage lambda, Curr Opin Genet Dev 15 (2005) 145-152.
[16] I.B. Dodd, A.J. Perkins, D. Tsemitsidis, J.B. Egan, Octamerization of lambda CI repressor is needed for effective repression of P(RM) and efficient switching from lysogeny, Genes Dev 15 (2001) 3013-3022.
[17] I.B. Dodd, K.E. Shearwin, A.J. Perkins, T. Burr, A. Hochschild, J.B. Egan, Cooperativity in long-range gene regulation by the lambda CI repressor, Genes Dev 18 (2004) 344-354.
[18] B. Revet, B. von Wilcken-Bergmann, H. Bessert, A. Barker, B. Muller-Hill, Four dimers of lambda repressor bound to two suitably spaced pairs of lambda operators form octamers and DNA loops over large distances, Curr Biol 9 (1999) 151-154.
[19] S.L. Svenningsen, N. Costantino, D.L. Court, S. Adhya, On the role of Cro in lambda prophage induction, Proc Natl Acad Sci U S A 102 (2005) 4465-4469.
[20] B.J. Meyer, M. Ptashne, Gene regulation at the right operator (OR) of bacteriophage lambda. III. lambda repressor directly activates gene transcription, J Mol Biol 139 (1980) 195-205.
[21] D. Jain, B.E. Nickels, L. Sun, A. Hochschild, S.A. Darst, Structure of a ternary transcription activation complex, Mol Cell 13 (2004) 45-53.
[22] B.E. Nickels, S.L. Dove, K.S. Murakami, S.A. Darst, A. Hochschild, Protein-protein and protein-DNA interactions of sigma70 region 4 involved in transcription activation by lambdacI, J Mol Biol 324 (2002) 17-34.
[23] M.E. Furth, W.F. Dove, B.J. Meyer, Specificity determinants for bacteriophage lambda DNA replication. III. Activation of replication in lambda ric mutants by transcription outside of ori, J Mol Biol 154 (1982) 65-83.
[24] K. Mensa-Wilmot, K. Carroll, R. McMacken, Transcriptional activation of bacteriophage lambda DNA replication in vitro: regulatory role of histone-like protein HU of Escherichia coli, EMBO J 8 (1989) 2393-2402.
[25] M.S. Wold, J.B. Mallory, J.D. Roberts, J.H. LeBowitz, R. McMacken, Initiation of bacteriophage lambda DNA replication in vitro with purified lambda replication proteins, Proc Natl Acad Sci U S A 79 (1982) 6176-6180.
[26] D.L. Court, A.B. Oppenheim, S.L. Adhya, A new look at bacteriophage lambda genetic networks, J Bacteriol 189 (2007) 298-304.
[27] C.E. Bell, M. Lewis, Crystal structure of the lambda repressor C-terminal domain octamer, J Mol Biol 314 (2001) 1127-1136.
[back]
oriT R ([http://partsregistry.org/wiki/index.php?title=Part:BBa_J01003 BBa_J01003])
We have characterized the oriT [http://partsregistry.org/wiki/index.php?title=Part:BBa_J01003 BBa_J01003] in a conjugation system in which pUB307 is used as helper plasmid and E. coli as donor and recipient cells. We measured quantitatively the conjugative competence of oriT [http://partsregistry.org/wiki/index.php?title=Part:BBa_J01003 BBa_J01003] by conjugation between different strains, at different temperatures and with different donor/recipient ratios. We also compared the conjugation kinetics of [http://partsregistry.org/wiki/index.php?title=Part:BBa_J01003 BBa_J01003] and pUB307 quantitatively.
The part oriT R [http://partsregistry.org/wiki/index.php?title=Part:BBa_J01003 BBa_J01003] was cotransformed together with pUB307 (helper plasmid) into E. coli strain Top10. These cells therefore became conjugation donor and have a kanamycin (from pUB307) and ampicillin (from [http://partsregistry.org/wiki/index.php?title=Part:BBa_J01003 BBa_J01003]) resistance. The recipient cells have the plasmid pBAD33 with a chloramphenicol resistance. After mixing both cell types the conjugation starts and [http://partsregistry.org/wiki/index.php?title=Part:BBa_J01003 BBa_J01003] as well as pUB307 can be transported. The helper plasmid pUB307 can still be transported since it still contains its oriT region. The transconjugants which have received [http://partsregistry.org/wiki/index.php?title=Part:BBa_J01003 BBa_J01003] through the conjugation have ampicillin and chloramphenicol resistance and can therefore be easily selected on agar plates containing these two antibiotics. [back]
Test 1 Conjugation kinetics of oriT [http://partsregistry.org/wiki/index.php?title=Part:BBa_J01003 BBa_J01003] between E. coli Top10
On the first day we inoculated LB + antibiotics medium with a single colony or from a glycerol stock. For this test we used donor and the recipient cells that were both E. coli Top10. On the second day we washed the cells of the overnight culture twice with LB medium, transferred the donor cell suspension to the pellet of the recipient cells and resuspended the recipient cells in this medium. After another centrifugation step we resuspended the pellet which now is a mixture of donor and recipient cells in 100 µl LB medium without antibiotics. We than transferred the cell suspension on a membrane filter, which was placed on LB plates before and incubated these plates at 37 °C for 0, 6, 12, 18, 24, 30, 36, 42 minutes. Afterwards the membrane was put into 1 ml LB medium and the cells were resuspended by vortexing for 30 seconds. The cell suspension was diluted 10-5 and plated on the LB / ampicillin + chloramphenicol (CA) plates. The plates were incubated at 37 °C overnight and on the third day the colonies could be counted.
We have done this test thrice independently and calculated the conjugation efficient which is defined as [transconjugants per ml] / [donor per ml] from this assay.
To count the donor and recipient cell density small aliquots of the used suspensions were diluted 10-7 and plated on agar plates containing kanamycin + ampicillin for donor cells and chloramphenicol for the recipients.
[back]
Table 1: Conjugation kinetics of oriT BBa_J01003 between E. coli Top10.
| Group | Test A | Test B | Test C | |||
| Cell | Donor | Recipient | Donor | Recipient | Donor | Recipient |
| Strain | Top10 | Top10 | Top10 | Top10 | Top10 | Top10 |
| Cell number | 9325 | 8100 | 3465 | 8330 | 7140 | 7470 |
| Time [min] | Transconjugants | Efficiency | Transconjugants | Efficiency | Transconjugants | Efficiency |
| 0 | 150 | 0.016 | 9 | 0,003 | 37 | 0,005 |
| 6 | 500 | 0.054 | 20 | 0,006 | 390 | 0,055 |
| 12 | 780 | 0.084 | 94 | 0,027 | 1390 | 0,195 |
| 18 | 1160 | 0.124 | 172 | 0,050 | 1720 | 0,241 |
| 24 | 1400 | 0.150 | 262 | 0,076 | 2880 | 0,403 |
| 30 | 3360 | 0.360 | 1450 | 0,418 | 3500 | 0,490 |
| 36 | 1790 | 0.517 | 3750 | 0,525 | ||
| 42 | 2170 | 0.626 | ||||
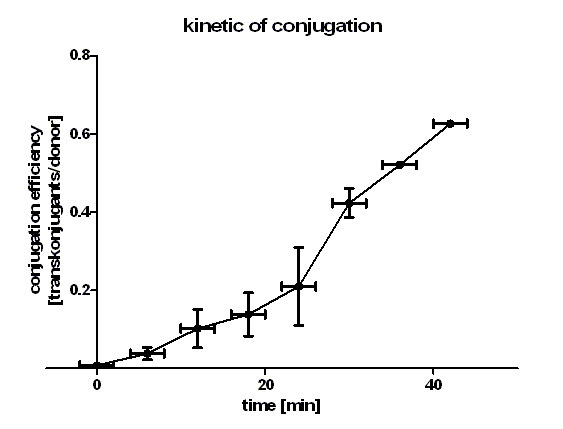
Figure 1 shows that the kinetic of the conjugative plasmid [http://partsregistry.org/wiki/index.php?title=Part:BBa_J01003 BBa_J01003] has three phases. In the first (0 min to 24 min) and in the third phase (30 min to 42 min) the curve has a smaller slope, whereas the second phase (24 min to 30 min) it has a very big slope. An explanation for this result could be that the cell growth in the first 24 min can be neglected due to the average generation time of E. coli in the LB medium which lies between 20 and 30 minutes whereas in the second phase the growth of E. coli dominates the conjugation process and leads to this deviation from linearity. In this test we used very concentrated bacteria suspension in a small conjugation volume which leads to the observation of conjugation in an approximately linear process in the primary phase.
We have done a linear regression analysis for the first phase (0 min to 24 min) and we calculated the slope for this phase as 9.00 • 10-3 [transconjugants • donor-1 • minute -1]. We calculated the conjugation rate for [http://partsregistry.org/wiki/index.php?title=Part:BBa_J01003 BBa_J01003] which is defined as transconjugants • donor-1 • recipient-1 • minute -1. The conjugation rate of [http://partsregistry.org/wiki/index.php?title=Part:BBa_J01003 BBa_J01003] is 1.05 • 10-12 [ml • min-1 • cell-1]. This rate was used by the modeling group as a parameter value.
Figure 1 also shows us that the transport of the plasmid pSB1A2 containing the part [http://partsregistry.org/wiki/index.php?title=Part:BBa_J01003 BBa_J01003] by conjugation is a very fast process since we got transconjugants in the first minutes of the experiment. Through this experiment we can conclude that the transport of the plasmid pSB1A2 containing [http://partsregistry.org/wiki/index.php?title=Part:BBa_J01003 BBa_J01003] by conjugation needs only about 2 minutes.
Since we have two conjugative plasmids in our donor cells, we wanted to see which plasmid is preferred to be transported by conjugation. We therefore plated the cells in the test C (see table 2) also on agar plates containing chloramphenicol + kanamycin (CK) and chloramphenicol + kanamycin + ampicillin (CKA) to select for transconjugants getting pUB307 and both plasmids.
[back]
Table 2: Test C conjugation kinetics of oriT BBa_J01003 and pUB307 between E. coli Top10.
| Select | Chlorophencol+Kanamycin (J01103) | Chlorophencol+Kanamycin (pUB307) | Chlorophenicol+Ampicilin+Kanamycin | |||
| Cell | Donor | Recipient | Donor | Recipient | Donor | Recipient |
| Strain | Top10 | Top10 | Top10 | Top10 | Top10 | Top10 |
| Cell number | 7140 | 7470 | 7140 | 7470 | 7140 | 7470 |
| Time [min] | Transconjugants | Efficiency | Transconjugants | Efficiency | Transconjugants | Efficiency |
| 0 | 37 | 0.005 | 2 | 0,000 | 1 | 0,000 |
| 6 | 390 | 0.055 | 17 | 0,002 | 4 | 0,001 |
| 12 | 1390 | 0.195 | 194 | 0,027 | 90 | 0,013 |
| 18 | 1720 | 0.241 | 230 | 0,032 | 109 | 0,015 |
| 24 | 2880 | 0.403 | 1130 | 0,158 | 800 | 0,112 |
| 30 | 3500 | 0.490 | 2500 | 0,350 | 1540 | 0,216 |
| 36 | 3750 | 0.525 | 2600 | 0,364 | 1800 | 0,252 |

Figure 2 clearly shows that the plasmid pSB1A2 containing only [http://partsregistry.org/wiki/index.php?title=Part:BBa_J01003 BBa_J01003] is preferably transported during the first 20 minutes in comparison to pUB307. Since pUB307 has the same oriT as [http://partsregistry.org/wiki/index.php?title=Part:BBa_J01003 BBa_J01003] is, we assume the size of the plasmid to be the reason for this since [http://partsregistry.org/wiki/index.php?title=Part:BBa_J01003 BBa_J01003] in pSB1A2 is approx. 2.5 kb in size whereas pUB307 has 60 kb. A comparison of the red and the green curve shows us that the difference in the duration of the transport of plasmids with different size is not very big. Looking at the blue curve you can see that the first transconjugants with both plasmids can be detected after approx. 18 minutes which is longer than the sum of both transport times.
[back]
Test 2 Conjugation kinetics of oriT [http://partsregistry.org/wiki/index.php?title=Part:BBa_J01003 BBa_J01003] between different E.coli strains
For this experiment we used the same technique as described in test 1. In this case we used different E. coli strains as recipients, namely E. coli Top10, MG1655 and DH5α. In this experiment the cells on the membranes were incubated for 0, 6, 12, 18, 24, 30, 36 and 42 minutes at 37 °C. This test was conducted twice independently and the conjugation efficiency calculated as described in test 1. [back]
Table 3: Conjugation kinetics of oriT BBa_J01004 between different E. coli strains.
| Group | Test D (Donor: Top10; Cell number: 5440) | Test E (Donor: Top10; Cell number: 7140) | ||||
| Cell | Recipient | Recipient | Recipient | Recipient | Recipient | Recipient |
| Strain | Top10 | MG1655 | DH5alpha | Top10 | MG1655 | DH5alpha |
| Cell number | 6086 | 5566 | 5096 | 7470 | 7275 | 6900 |
| Time [min] | Efficiency | Efficiency | Efficiency | Efficiency | Efficiency | Efficiency |
| 0 | 0.002 | 0,000 | 0,000 | 0,005 | 0,000 | 0,001 |
| 6 | 0.002 | 0,000 | 0,000 | 0,055 | 0,000 | 0,003 |
| 12 | 0.003 | 0,000 | 0,000 | 0,195 | 0,001 | 0,067 |
| 18 | 0.005 | 0,000 | 0,001 | 0,241 | 0,007 | 0,165 |
| 24 | 0.021 | 0,000 | 0,018 | 0,403 | 0,019 | 0,269 |
| 30 | 0.071 | 0,001 | 0,075 | 0,490 | 0,036 | 0,415 |
| 36 | 0.136 | 0,013 | 0,246 | 0,525 | 0,129 | 0,482 |
| 42 | 0.041 | 0,301 | 0,134 | 0,566 | ||
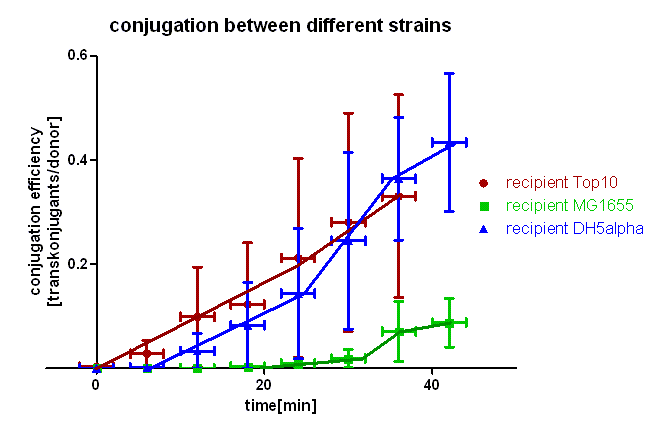
As you can see in figure 3 there is a difference in the conjugation kinetic between Top10 as donor and different strains as recipient. With E. coli Top10 as donor E. coli DH5α is a comparably good recipient as Top10 whereas MG1655 showed up as a bad recipient under these conditions. Comparing the red with the blue curve one can see that DH5α transconjugants appear later (6 minutes) that Top10, but the conjugation efficiency and also the conjugation rate (slope between 6 and 24 minutes) are in the same range with Top10 and DH5α. We assume that more time is needed for stabilizing the pillus contact between Top10 and DH5α than between two Top10 cells, but the conjugation between Top10 and DH5α can be done as good as between two Top10 cells. Comparing the red and the green curve one can see that the first MG1655 transconjugants appear much later (20 minutes) and the conjugation efficiency and rate (slope between 20 and 32 minutes) are much smaller. Therefore we conclude that conjugation between different strains is possible using this oriT but the efficiency depends on the used strains and their compatibility.
[back]
Test 3 Conjugative competence of oriT [http://partsregistry.org/wiki/index.php?title=Part:BBa_J01003 BBa_J01003] at different donor/recipient ratios
In this experiment we again used the same protocol as described above in test 1 but used different donor / recipient rations, namely 50:1, 25:1, 10:1, 8:1, 5:1, 3:1, 2:1, 1:1, 1:2, 1:3, 1:5, 1:10, 1:25, and 1:50. This test was conducted twice independently and the conjugation efficiency calculated as described above. The donor and recipient cell density was measured by plating 10-7 dilutions on agar plates containing kanamycin + ampicillin for the donor and chloramphenicol for recipient cells. [back]
Table 4: Conjugative competence of otiT BBa_J01003 at different donor / recipient ratios.
| Group | Test F | Test G | ||||||
| Anticipant ratio | Donor | Recipient | Actual ratio | Transconjugants | Donor | Recipient | Actual ratio | Transconjugants |
| 50:1 | 1003 | 23 | 43:1 | 12 | 832 | 21 | 40:1 | 16 |
| 25:1 | 982 | 46 | 21.5:1 | 28 | 815 | 41 | 20:1 | 31 |
| 10:1 | 930 | 110 | 8.5:1 | 62 | 729 | 97 | 7.5:1 | 63 |
| 8:1 | 909 | 133 | 7:1 | 81 | 711 | 117 | 6:1 | 75 |
| 5:1 | 854 | 202 | 4:1 | 115 | 665 | 153 | 4.5:1 | 120 |
| 3:1 | 767 | 302 | 2.5:1 | 161 | 599 | 266 | 2.3:1 | 129 |
| 2:1 | 680 | 403 | 1.7:1 | 213 | 533 | 355 | 1.5:1 | 212 |
| 1:1 | 510 | 605 | 1:1.2 | 302 | 400 | 532 | 1:1.3 | 303 |
| 1:2 | 340 | 802 | 1:2.3 | 351 | 268 | 705 | 1:2.6 | 298 |
| 1:3 | 257 | 902 | 1:3.5 | 294 | 199 | 794 | 1:4 | 302 |
| 1:5 | 170 | 1003 | 1:5.9 | 289 | 133 | 883 | 1:6.7 | 194 |
| 1:8 | 122 | 1072 | 1:8.8 | 241 | 89 | 943 | 1:10.5 | 190 |
| 1:10 | 94 | 1095 | 1:11.7 | 158 | 72 | 963 | 1:13 | 134 |
| 1:25 | 40 | 1159 | 1:29 | 86 | 31 | 1020 | 1:33 | 82 |
| 1:50 | 20 | 1182 | 1:59 | 31 | 16 | 1040 | 1:67 | 30 |

Figure 4 shows very clearly that at same cell density, if donor : recipient ratio is 1 : 2 the highest amount of transconjugants can be achieved. A big difference in the amount of donor : recipient leads to a low number of transconjugants. At low donor : recipient ratios the conjugation efficiency was quiet high but lead to a low number of transconjugants since the density of donor cells was too low. Only at donor : recipient rations between 1 : 1 and 1 : 3, a high conjugation efficiency and a high donor density was achieved leading to a high number of transconjugants. A maximum in the number of transconjugants could be achieved at a ratio of 1 : 2.
[back]
Test 4 Conjugative competence of oriT [http://partsregistry.org/wiki/index.php?title=Part:BBa_J01003 BBa_J01003] at different temperatures
In this experiment the same method is used as described above in test 1. Here, donor and recipient both were E. coli Top10. The incubation during conjugation took place at different temperatures, namely 4 °C, 22 °C, 30 °C, 37 °C and 42 °C for 20, 40 and 60 minutes. For counting of transconjugants the cell suspension was diluted 10-6 and plated on agar containing ampicillin and chloramphenicol (CA). This experiment was conducted twice independently. The donor and recipient cell density was again calculated by plating 10-7 dilutions on plates containing kanamycin and ampicillin or chloramphenicol, respectively. [back]
Table 5: Conjugative competence of otiT BBa_J01003 at different temperatures.
| Group | Test H | Test J | ||||
| Cell | Donor | Recipient | donor : recipient | Donor | Recipient | donor/recipient |
| Strain | Top10 | Top10 | 1 : 2.02 | Top10 | Top10 | 1:1.77 |
| Cell number | 361 | 731 | sum:1092 | 330 | 584 | sum:914 |
| Sample | Transconjugants | Transconjugants | ||||
| Temperature | 10min | 40min | 60min | 20min | 40min | 60min |
| 4°C | 5 | 5 | 1 | 2 | 1 | 0 |
| 22°C | 2 | 78 | 117 | 29 | 61 | 79 |
| 30°C | 71 | 178 | 241 | 43 | 117 | 227 |
| 37°C | 154 | 285 | 550 | 118 | 290 | 332 |
| 42°C | 167 | 273 | 402 | 122 | 271 | 234 |
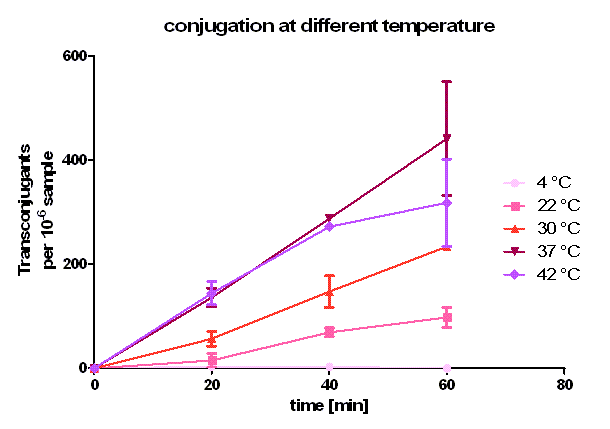
As you can see in figure 5, conjugation is most efficient at 37 °C. From 0 to 20 minutes during the primary conjugation phase the cell growth can be neglected. From 20 to 40 minutes, the cell growth is also presented in the conjugation curves. At 4 °C, the conjugation curve has a very small slope (near zero) and no significant transconjugants production could be observed. At 22, 30, 37 and 42 °C, a significant number of transconjugants could be achieved, but the conjugation efficiency and the conjugation rate (slope between 0 and 20 minutes) differ strongly. From 22 °C to 37°C, the conjugation efficiency and conjugation rate increases with rising temperature. Comparing these three curves between 20 and 40 minutes one can see that the cell growth gets more important with rising temperature. Dynamically, the cells have more mobility at higher temperature, and have a bigger possibility to touch another cell, which is necessary for conjugation. When we compared the conjugation curve at 42 °C with the one at 37 °C, one can see, that the conjugation efficient and the conjugation rate at 42 °C is as good as at 37 °C. After the primary conjugation phase, the production of transconjugants at 42 °C is clearly worse than the production at 37 °C what can be seen at comparing the curves between 40 and 60 minutes. A possible reason for this could be a lower growth and higher death rate at 42 °C compared to 37 °C. So, one can conclude, that for best conjugation efficiency one needs a warm environment for the conjugation to take place but also temperatures which favor cell growth.
[back]
Summary
The characterization of [http://partsregistry.org/wiki/index.php?title=Part:BBa_J01003 BBa_J01003] in pSB1A2 has given the following results:
- The conjugation has an average conjugation rate of 1.05 • 10-12 [ml • min-1 • cell-1]
- The conjugation has an approximate transport time of 2 minutes
- The oriT [http://partsregistry.org/wiki/index.php?title=Part:BBa_J01003 BBa_J01003] will be transported first in attendance of pUB307 in the donor cells
- Smaller plasmids favor the conjugation and are conjugated first
- The oriT [http://partsregistry.org/wiki/index.php?title=Part:BBa_J01003 BBa_J01003] can be conjugated between Top10 cells and from Top10 to DH5α cells with similar efficiency but from Top10 to MG1655 with a strongly lower efficiency
- The conjugation favors a donor : recipient ratio of 1 : 2
- The conjugation has the best production of transconjugants, the best efficiency and the best conjugation rate at 37 °C
[back]
Chloramphenicol resistance cassette ([http://partsregistry.org/wiki/index.php?title=Part:BBa_K150003 BBa_K150003])
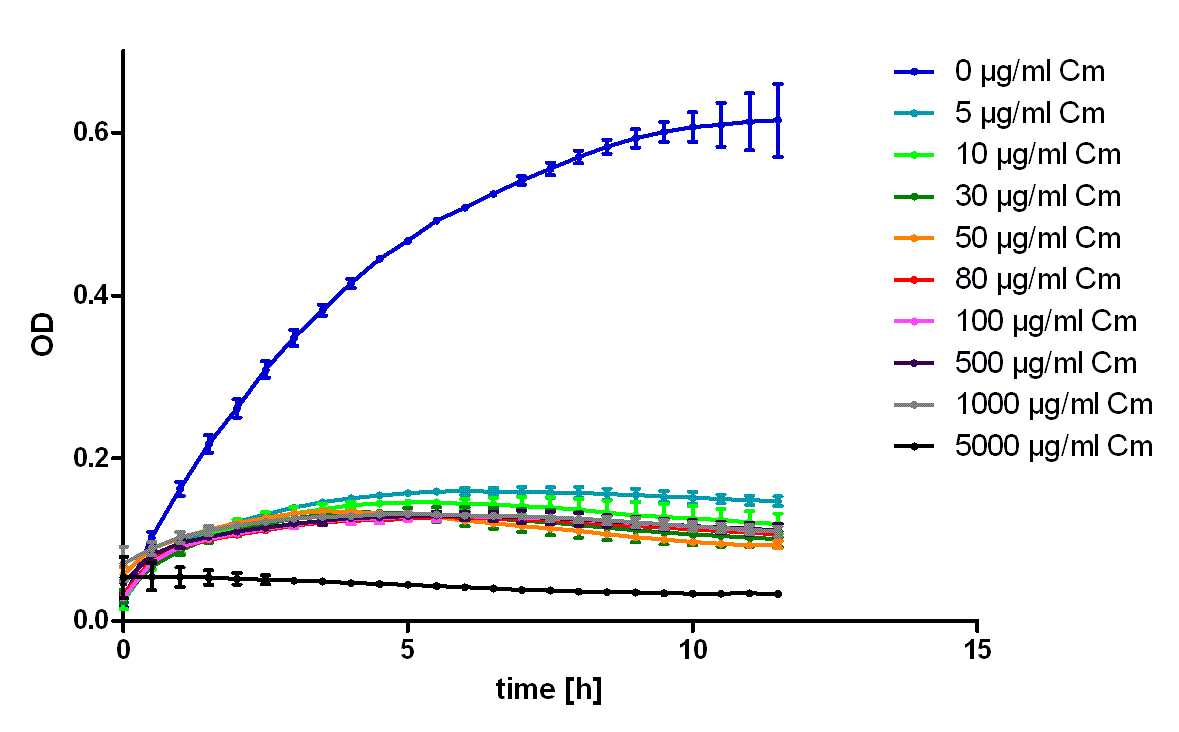
To characterize the ability of this protein generator to mediate resistance against chloramphenicol we used E. coli Top10 transformed with the part BBa_K150003 and measured their ability to grow in LB medium containing different amounts of chloramphenicol (see figure 2). As control we used untransformed E. coli Top10 (see figure 1). To exclude the possibility that high concentrations of chloramphenicol led to the death of the bacteria only through the solvent ethanol we also measured the same ethanol concentrations as mock treated (see figure 3).
It was shown that this part leads to acceptable resistance up to 100 µg/ml chloramphenicol, even in 500 µg/ml unmistakably cell growth can be detected. The effect of the solvent ethanol can even be at high concentrations neglected.
LuxR-Colicin E1-Receiver ([http://partsregistry.org/wiki/index.php?title=Part:BBa_K150009 BBa_K150009])
ColicinE1 Producer Controlled by 3OC6HSL Receiver Device - Introduction
The constitutive expressed LuxR protein is able to build a complex with an AHL molecule. [7] This complex functions as transcription factor for the lux pR promoter and activates therefore colicinE1 production. The activation of the lux pR promoter leads additionally to expression of the kil protein, also called lysis protein by readthrough of a terminator located in the colicinE1 operon. Production of the lysis protein leads to lysis of the host cell and colicin release. The colicinE1 immunity protein (imm) located on the opposite strand is expressed under a constitutive promoter and protects the host cell from lysis up to a certain threshold of the lysis protein. [back]
Colicin E1 - Mode of Action
Colicin E1 is a 522 residue containing protein and belongs to the group A colicins. It kills sensitive cells, which harbor no ColE1 plasmid but contain the appropriated receptor BtuB, by forming pores, or more precisely ion channels into the inner membrane. [1] Affected cells can be E. colis or related strains but also some eukaryotic cells have been affected by the toxic activity of colicin. [2] The first step of the lethal action of colicin E1 is binding to the vitamin B12 receptor (BtuB). Colicins are able to kill bacteria by a “one hit” mechanism. This works due to destruction of the target cells cellular energy. The voltage dependant channel in the cytoplasmatic membrane leads to a depolarization of the target cell. The conductance of the channel has been shown to be ~107 ions/channel-sec in 1 M NaCl. [3] Its ability to form pores is given by the C-terminal domain. This domain consists of 10 tightly packed α–helices (see Figure 1). [4], [6] The colicin protein is a water soluble molecule containing mostly amphipathic helices except from two hydrophobic ones. Due to these two hydrophobic helices the protein is able to target the cell membrane and act as a membrane protein. [back]
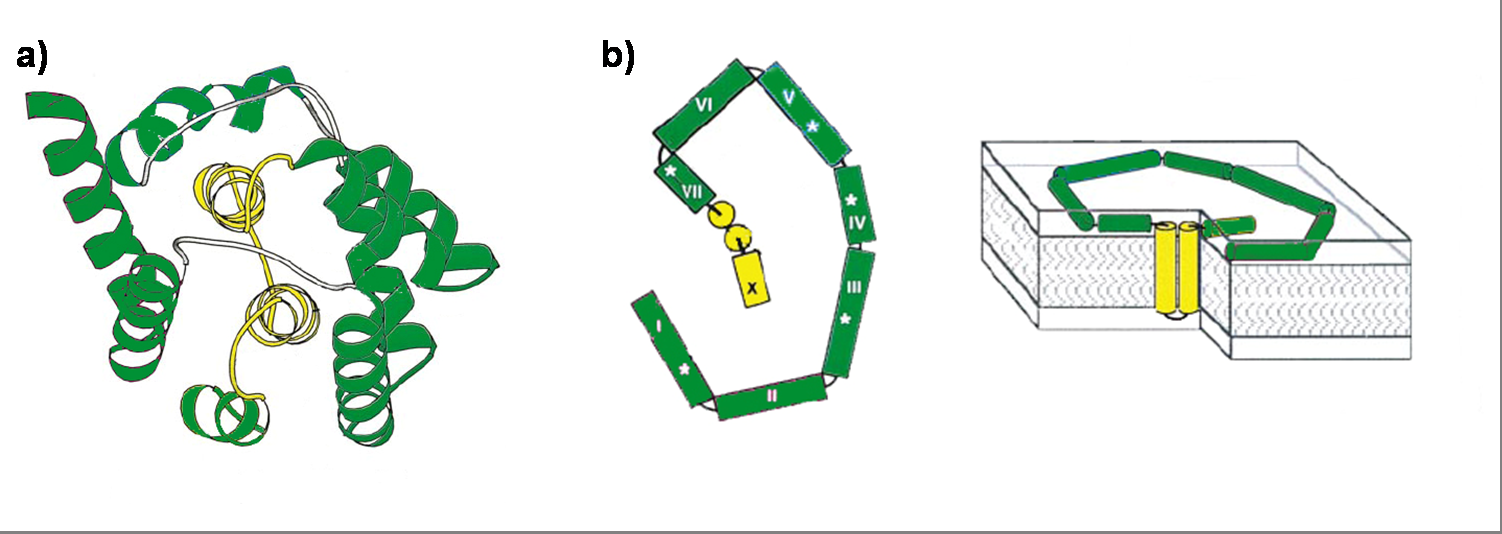
Killing efficiency on bacteria
To measure the killing efficiency and which amount of cells or colicins are needed to reach any killing activity a colicin activity test was carried out. Therefore bacteria containing the colicin part ([http://partsregistry.org/wiki/index.php?title=Part:BBa_K150009 BBa_K150009] in TOP10 or MG1655) and GFP producing cells (reference promoter, TOP10) were inoculated in TB-media with appropriate antibiotics at 37 °C for 4 to 6 hours and the optical density of the two strains was adjusted. The colicin cells were added in different ratios to a constant amount of GFP producing cells. The total volume was kept constant by adding TB-media (without antibiotics). The colicin production was induced by several concentrations (0 M-100 nM) of N-Acyl-Homoserin-Lactone (AHL). The OD and GFP intensities were measured at 37 °C in the Tecan Microplate Reader every 30 minutes for about 12 hours. As a negative control the similar test was carried out with cells containing the same plasmid without the colicin gene on it. Figure 2 shows results of tests with a prey-killer ratio of 100:1. Due to the correlation of the GFP intensity and the optical density, GFP intensity was used as marker for the living prey cells. In all experiments using killer cells and high AHL concentrations in the medium, the prey cells were killed completely. In reference experiments (see Figure 2, bottom) using E. coli TOP 10 cells harboring a LuxR-receiver without colicin operon (comparable to part [http://partsregistry.org/wiki/index.php?title=Part:BBa_T9002 BBa_T9002] without GFP), the prey cells were able to grow for each AHL concentration. Consequently the lethal action of the killing part could be shown. [back]
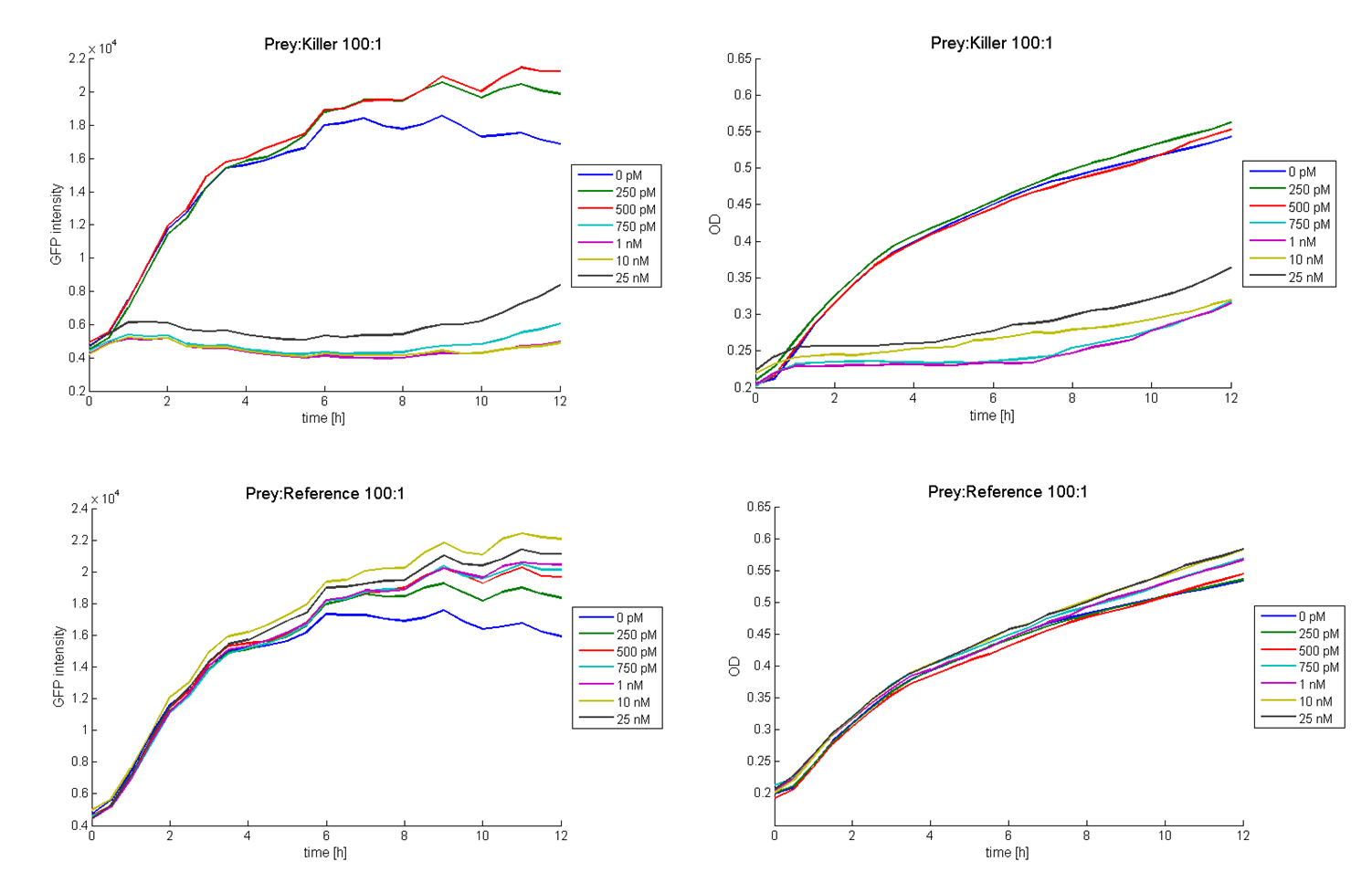
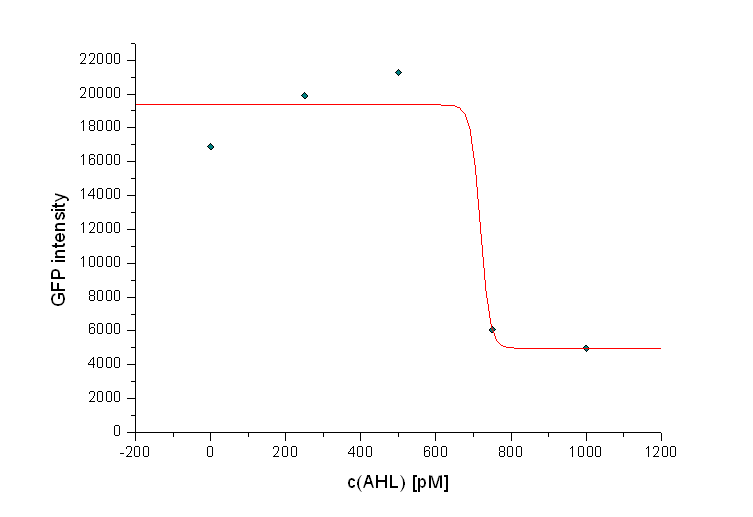
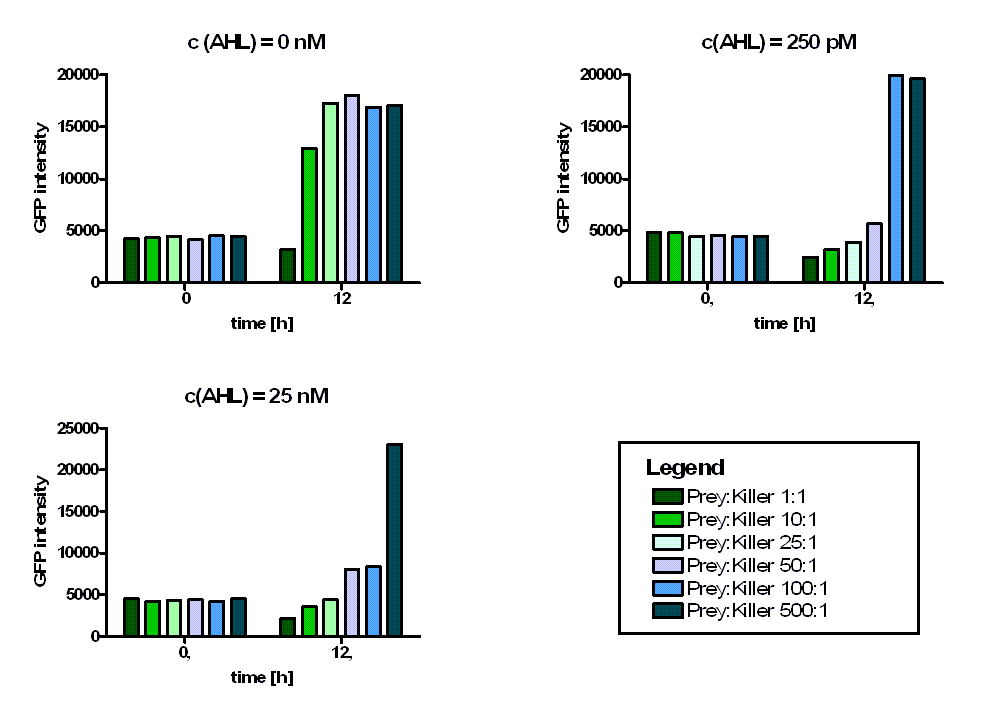
Additionally it can be seen that the killing efficiency is related to the AHL concentration in the medium. Figure 3 shows a dose-response curve of GFP intensity after 12 hours dependent on the AHL concentration. The PLuxR promoter has to be activated with an AHL concentration above 500 pM. Thus enough colicin E1 is produced and released to kill the prey cells (see Figure 3).
[back]
Regarding the prey survival, dependent on the prey-killer ratio, several effects can be observed (see Figure 4). Having a prey-killer ratio of 1:1, or even a higher killer fraction, all prey cells were already killed when no AHL is present. This effect is caused by the leakiness of the PLuxR promoter. For prey-killer ratios between 5:1 and 100:1 the killing efficiency can be regulated by the AHL concentration. In experiments with prey-killer ratios higher than 100:1 the growth of the prey strain was not influenced.
[back]
Killer-prey system
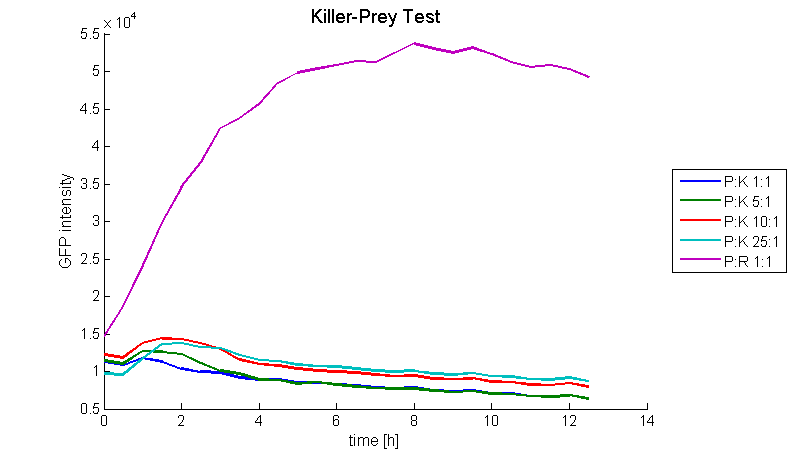
To measure the functionality the killer-prey system were tested. Therefore bacteria containing the colicin E1 part ([http://partsregistry.org/wiki/index.php?title=Part:BBa_K150009 BBa_K150009] in TOP10 or MG1655) and prey cells containing an AHL-producing part ([http://partsregistry.org/wiki/index.php?title=Part:BBa_K150000 BBa_K150000]) were inoculated in TB-media with appropriate antibiotics at 37 °C for 4 to 6 hours. The optical density of the two strains was adjusted. The colicin E1 producing cells were added in different ratios to a constant amount of prey cells. The total volume was kept constant by adding TB-media (without antibiotics). The colicin production was induced by several concentrations (0 M-100 nM) of N-Acyl-Homoserin-Lactone (AHL). The OD and GFP intensities were measured at 37 °C in the Tecan Microplate Reader every 30 minutes for about 12 hours. As a negative control the similar test was carried out with cells, containing the same plasmid without the colicin E1 gene on it. Figure 5 shows that the prey cells were killed for prey-killer ratios from 1:1 up to 25:1. Therefore it is proven that our system works as expected. [back]
Lysis of killing strain
Besides the killing efficiency the time course of the killer strain lysis was analyzed. Therefore growth curves of killer cells ([http://partsregistry.org/wiki/index.php?title=Part:BBa_K150009 BBa_K150009] in E. coli TOP10 cells) induced with different AHL concentrations were measured (see Figure 6).
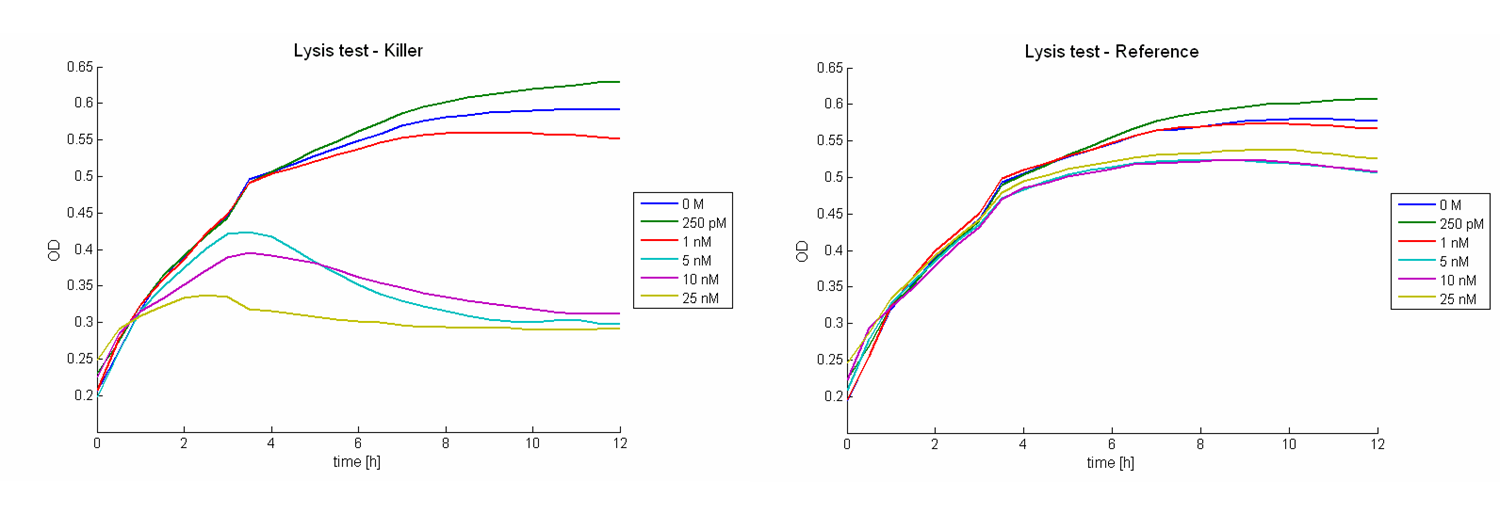
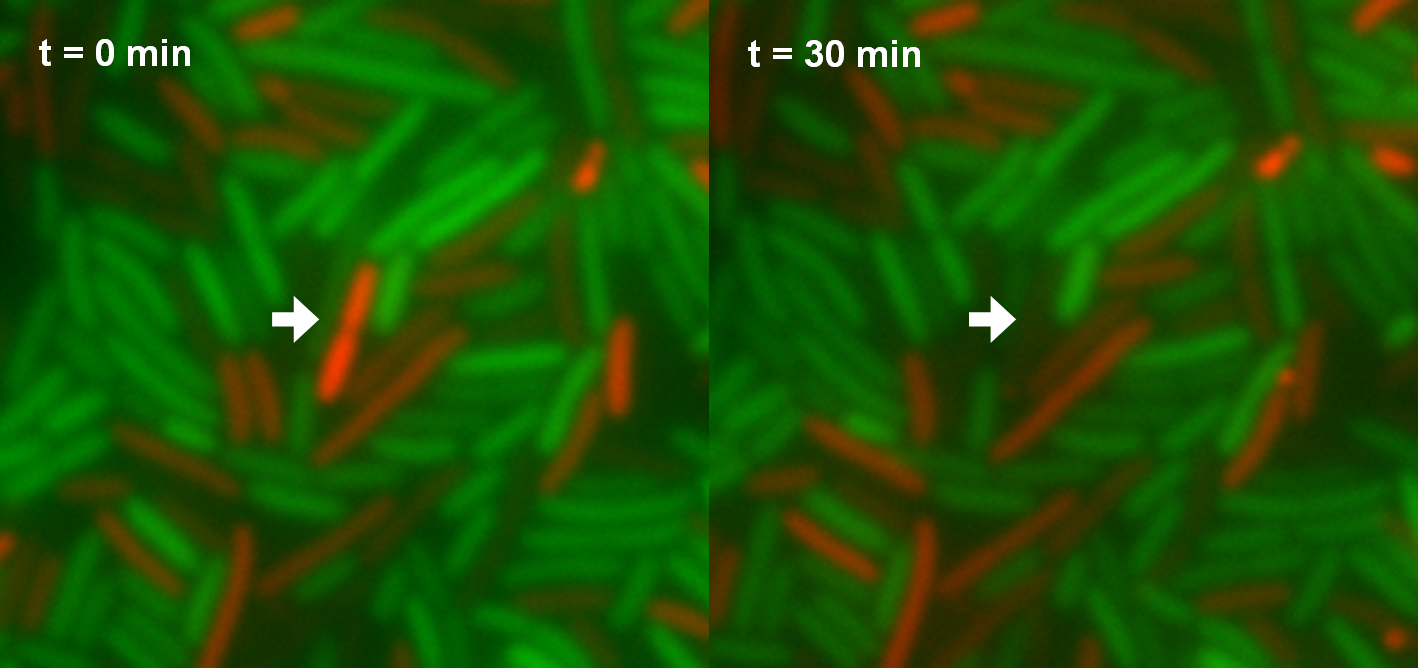
During the first hour of the measurement no effect can be observed. Afterwards cell lysis dependent on AHL concentrations is visible. For AHL concentrations between 0 M and 1 nM only few cells lysed. Thus growth of the population is hardly influenced. Concentrations from 5 nM up to 25 nM show a strong effect on the amount of lysed killer cells. For 5 nM and 10 nM the growth curves flatten out. After reaching a maximum during the third and fourth hour the population size decreases and converges to a constant value. For higher AHL concentrations, e.g. 25 nM, a similar effect can be observed, but it is much stronger. The population shows only a weak growing tendency but then converges directly to a constant value. In addition to these growth measurements lysis of the killer cells was observed under the microscope. Figure 7 shows killer cells which were induced by AHL (left panel, t = 0 min). After 30 minutes one cell is lysed (right panel).
[back]
Eukaryotic killing Assay (Preliminary Results)
The toxicity of colicin E1 on eukaryotic cells was tested by using breast cancer cells (MCF 7). They were inoculated with killer cells (BBa_K150009 in TOP 10) and reference cells (BBa_150009-like, no colicin E1 operon) for several hours.
Therefore MCF-7 cells (breast cancer cell line) were plated in 24 well plates (150 thousand cells per well) with 10 % FCS (fetal calf serum) DMEM (Dulbecco's Modified Eagle Medium) and incubated overnight at 37 °C, 5 % CO2. E. coli TOP 10 strains containing LuxR-colicin E1-receiver (BBa_K150009) and strains containing LuxR-receiver (no colicin) were grown overnight in TB media with appropriate antibiotics at 37 °C to an OD of 1.0 and diluted in the morning to an OD of 0.1. Once bacterial cells reached an OD of 0.35 they were harvested and inoculated on the MCF-7 wt and on MCF-7 GFP producing cells. [back]
Once MCF-7 cells were confluent (12 h after passage), E. coli TOP 10 bacteria were spinned down (1 ml of OD = 0.35) and resuspended with DMEM medium from the original wells containing the MCF-7 wt/MCF-7 GFP cells. A 24-well plate was used containing MCF-7 cells stained with Propidium Iodide (PI) alone, with LuxR-receiver cells not containing colicin and LuxR-receiver-colicin E1 cells. AHL was added to every well to a final concentration of 5 nM. Well content was collected at different time-points (0 – 6 h) and when necessary PI was added to the suspension 15 minutes before Flow Cytometer analysis.[back]
At the referred timepoints, supernatants were collected and spinned down. Eukaryotic cells were detached by trypsinization (200 μl) for 10 minutes at 37 °C and then resuspended in 800 μl of KH + 1 % BSA (Krebs Henseleit medium with 1 % Bovine Serum Albumin). After resuspending, well content was added to the precipitated supernatant to account for floating dead cells. PI was added to each sample (1:1000) and incubated at RT for 15 minutes before acquisition. PI positive cells and GFP expressing cells (%) were measured for 3000 cells for each condition. Histograms and fluorescent measurements were obtained using a Cytomics FC 500 MPL from Beckman&Coulter. [back]
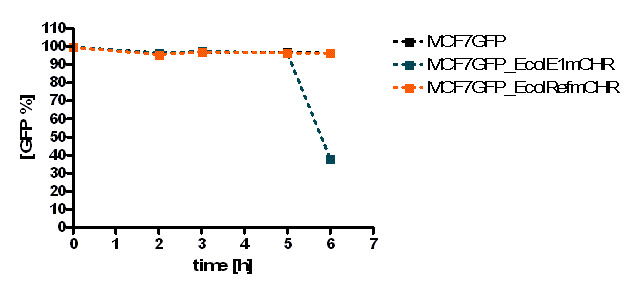
MCF-7 cell death, measured by Flow Cytometry, was evaluated by % of stained cells with PI over the time, using colicin producing E. coli or strains which do not produce colicins as a control (Figure 1). E. coli was also quantified (data not shown). The PI measurements (filter FL3-620nm) were gated only to Mcf-7 cell population. [back]
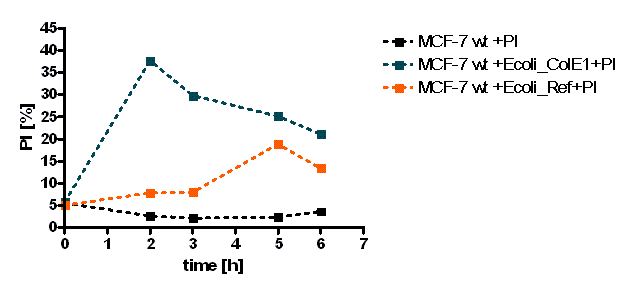
[back]
The initial results show that killing by E. coli producing colicin E1 is effective on eukaryotic cancer cells (MCF-7). Propidium iodide (or PI) is an intercalating agent and a fluorescent molecule with a molecular mass of 668.4 Da that can be used to stain DNA. It can be used to differentiate, apoptotic and normal cells, since it is membrane impermeant and generally excluded from viable cells. The high percentage of positive PI cells refers to cells that are permeable to it, thus, going into apoptosis. In this assay, after 2 h of incubation with the eukaryotic cells, the death rate was close to 40 % of the whole eukaryotic cell population. In comparison with the control conditions (MCF-7 alone + PI), where cells (maximum of 5%) due to trypsin treatment die, the effect is clearly visible. Furthermore, looking at the eukaryotic cell death after incubation with the reference strain (LuxR-receiver TOP 10), it can be suggested that the simple incubation of these mammalian cells with bacteria is causing cell death over time (maximum of 20% after 5 h). This effect is highly enhanced and faster when the bacteria produce colicin E1 (maximum of 37 % at 2 h). After 5 h, both bacterial strains produce similar results in killing the mammalian cancer cells. After reaching a peak, the killing efficiency drops, with a significant higher peak for E. coli bacteria producing colicin E1 than for the reference strain. On the other hand, using a stable cell line of MCF-7 expressing GFP, the effects of bacteria were not visible for the initial timepoints. The aim of this assay was to see the GFP signal droping down as E. coli progressed to kill the eukaryotic cells. Since GFP is a very robust protein we can suggest that colicin E1 producing bacteria start apoptotic events at early time (2 h, see Figure 8), but GFP loss can only be observed after a certain time delay (see Figure 9). For that reason, it would be necessary to observe also later timepoints to characterize the killing effect by GFP loss. [back]
Summerized, the preliminary results suggest that colicin E1 producing E. coli bacteria might be able to kill cancer cells at a high rate. However, further studies need to provide more insight into these events, as it might be by other pro-apoptotic markers or by repeating this experiment using different conditions (timepoints, autoinducer concentrations, another eukaryotic cell type). Future studies should yield to information about the colicin E1 target in eukaryotic cells and perhaps answers how it can kill cancer cells and spare healthy ones. [back]
References
[1] Cramer W. et al. (1990) Structure and dynamics of the colicin E1 channel. Molecular Microbiology 4: 519-526.
[2] Smarda J. et al. (2001) Cytotoxic effects of colicins E1 and E3 on v-myb-transformed chicken monoblasts. Folia Microbiol. (Praha) 47: 11–13.
[3] Bullock J. et al. (1983) Comparison of the macroscopic and single channel conductance properties of colicin E1 and its COOH-terminal tryptic peptide. J. Biol. Chem. 258: 9908–9912.
[4] Elkins P. et al. (1997) A mechanism for toxin insertion into membranes is suggested by the crystal structure of the channel-forming domains of Colicin E1. Structure. 5: 443-458.
[5] Lindeberg M. et al. (2000) Unfolding Pathway of the Colicin E1 Channel Protein on a Membrane Surface. J. Mol. Biol. 295: 679-692
[6] Davies, J. et al. (1975) Genetics of resistance to colicins in Escherichia coli K12: cross-resistance among resistance of group A. J. Bacteriol. 123:102–117
[7] Engebrecht J. et al. (1984) Identification of genes and gene products necessary for bacterial bioluminescence. Proc. Natl. Acad. Sci. USA 81: 4154–58
[back]
LuxI 3OC6HSL sender device ([http://partsregistry.org/wiki/index.php?title=Part:BBa_K150000 BBa_K150000])
General Description
AHL, a cell-cell signaling molecule is produced by the LuxI protein. The LuxI protein is expressed due to the luxI gene which is under the control of a constitutive promoter ([http://partsregistry.org/wiki/index.php?title=Part:BBa_J23107 BBa_J23107]). For this reason no input is needed for induction of gene expression. As output AHL can be measured. [back]
Quantification of AHL production
For AHL quantification the amount of AHL in supernatant of producing cells (E. coli TOP 10 containing [http://partsregistry.org/wiki/index.php?title=Part:BBa_K150000 BBa_K150000]) was determined. Therefore eight samples containing TB-media were inoculated with 2 µl/ml of an AHL producing cell overnight culture at 37 °C. Sterile filtered supernatant of these samples was produced hourly for 7 hours and stored at 4 °C. To estimate the amount of AHL producing cells the optical density was measured before. (Figure 1, top left). [back]
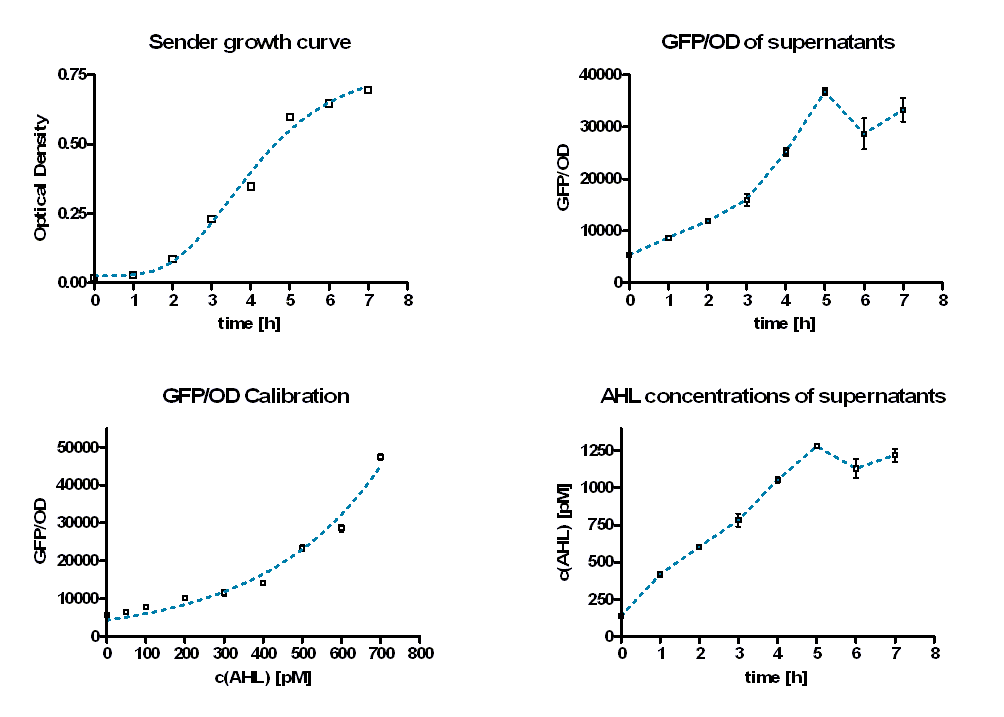
For the AHL determination 1 ml supernatant was diluted with 1 ml TB-media and inoculated with a constant amount of AHL inducible cells which produce GFP after induction ([http://partsregistry.org/wiki/index.php?title=Part:BBa_T9002 BBa_T9002] in E. coli TOP 10). The OD and GFP intensities were measured at 37 °C half-hourly for about 28 hours. For calibration the same test were performed with distinct AHL concentrations from 0M to 50 nM (SIGMA). Figure 2 shows the GFP/OD development over the time depending on the sender supernatants (left panel) or the preassigned AHL concentrations (right panel). The values increase and reach a maximum after 6 hours. Then they decrease slightly.
[back]
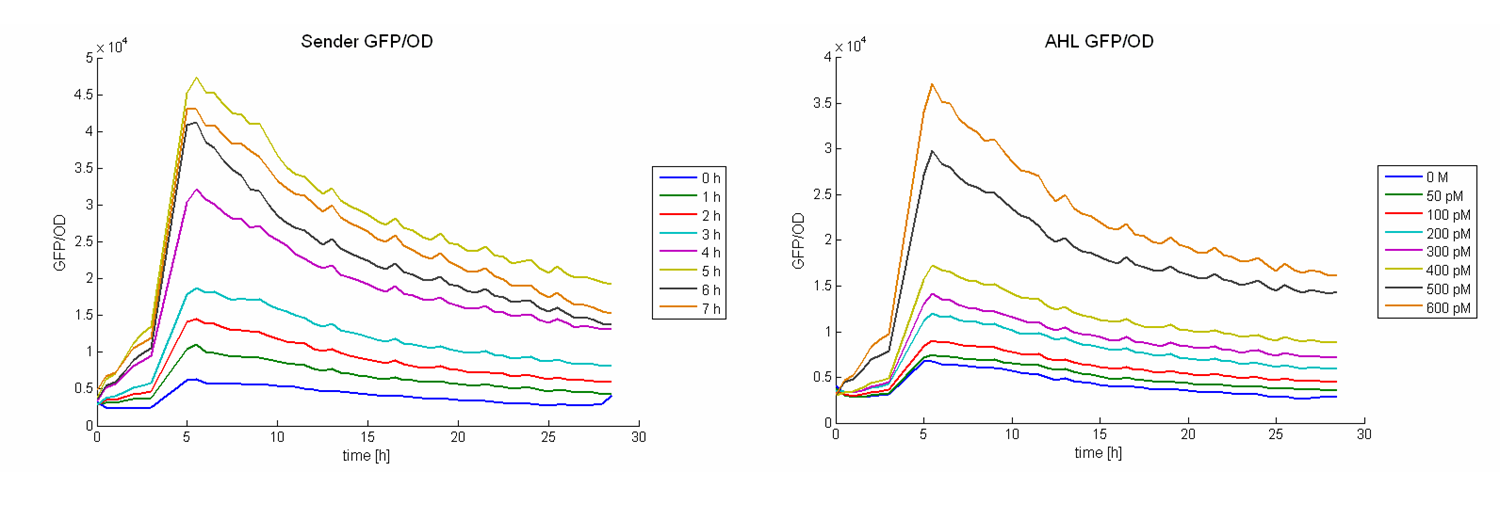
A GFP/OD – AHL calibration curve (Figure 1 bottom left) was calculated using the GFP/OD values at t = 10 h. An exponential growth curve was fitted to these data (blue dotted line; y = 4325 * e0.00346*x). The AHL concentrations of the sender supernatants were determined using the GFP/OD values at t = 10 h and the exponential regression. In the course of the time the AHL concentration increase until it reaches a certain maximum before a slight decrease (see Figure 1, bottom right, Table 1). The maximum is reached at the end of the logarithmic phase. Several tests have shown that the maximal concentration that can be produced by part [http://partsregistry.org/wiki/index.php?title=Part:BBa_K150000 BBa_K150000], is approximately 1100 and 1300 pM.
[back]
LuxS from Vibrio harveyi (partly characterized)
LuxS encodes for the protein (LuxS autoinducer synthase) that is responsible for the synthesis of autoinducer 2 (AI-2) in many species of Gram-negative and Gram-positive bacteria. This LuxS construct is cloned from Vibrio harveyi, a Gram-negative bioluminescent marine bacterium, which regulates light production in response to two distinct autoinducers (AI-1 and AI-2). [1] [back]
Functional check of LuxS/AI-2 production
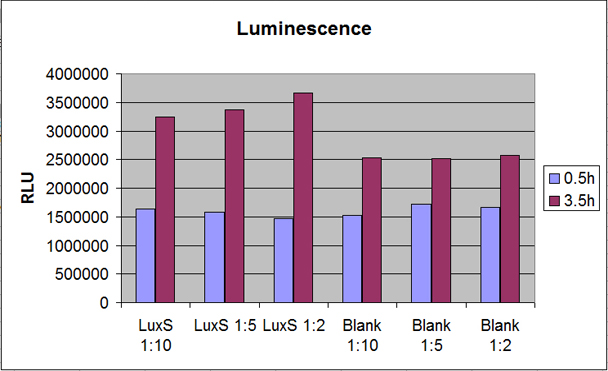
The functionality of the new LuxS part that was cloned by PCR from V. harveyi (BB120) in a ptrc99α plasmid was proven by measurement of luciferase activity in Vibrio as a reporter strain. DH5α cells with the new construct pTrc99α_LuxS were grown up and the supernatant was harvested by using centrifugation and sterile filtration. The gained supernatant was added to the reporter cells in different dilutions and incubated for three hours. Luminescence and OD were measured after 0.5h and 3.5h. [back]
Results
Cell growth rate of all samples LuxS and Blank was shown to be similar in OD measurements (data not shown). Luminescence of Vibrio reporter strain (data shown in figure 1) seems to increase with the concentration of supernatant in the cell delusion. Blanks with supernatant of DH5α cells transformed with pTrc99α plasmid show a significantly smaller Luminescence and there is no increase of Luminescence for increasing supernatant concentration in the cell delusion. [back]
Reference
[1] The LuxS family of bacterial autoinducers: biosynthesis of a novel quorum-sensing signal molecule. Mol Microbiol. 2001 Jul; 41(2):463-76.
[back]
 "
"