Team:BCCS-Bristol/Modeling-Agent Based
From 2008.igem.org
Tgorochowski (Talk | contribs) (→Framework Design) |
Tgorochowski (Talk | contribs) (→Simulation Process) |
||
| (8 intermediate revisions not shown) | |||
| Line 22: | Line 22: | ||
The stochastic agent based simulation framework we have developed is called BSim. This allows for differing configurations of particles and bacteria to easily be modelled in an interactive way. The framework has been developed from scratch in Java allowing it to be run on the majority of computer systems. In addition to an easy to use GUI, a command line based interface is also provided to permit the running of large scale simulations on remote computing clusters. | The stochastic agent based simulation framework we have developed is called BSim. This allows for differing configurations of particles and bacteria to easily be modelled in an interactive way. The framework has been developed from scratch in Java allowing it to be run on the majority of computer systems. In addition to an easy to use GUI, a command line based interface is also provided to permit the running of large scale simulations on remote computing clusters. | ||
| - | === | + | === Framework Design === |
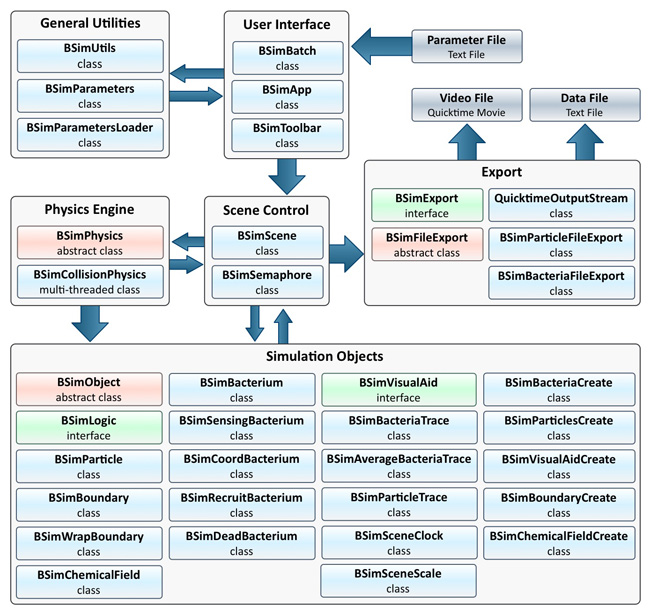
| + | [[Image:BCCS-Modelling_BSim_Classes.jpg|center|thumb|400px|'''Figure 2.5:''' BSim Class Diagram]] | ||
| + | |||
| + | The BSim framework is a cross platform Java based programming API that has been developed | ||
| + | from scratch to model the stochastic interactions of bacteria and particles in a fluid based environment. | ||
| + | Taking this approach allowed for the greatest flexibility including features essential to | ||
| + | our models. While this lead to an initially large effort in getting a working system completed, | ||
| + | it later significantly increased the speed at which additional aspects could be incorporated. | ||
| + | BSim has been developed using an object-oriented methodology and split into several highlevel | ||
| + | modules shown in Figure 2.5. To give end-users the option of running the system on | ||
| + | headless computers, e.g. via remote terminals, two alternative interfaces were created. The | ||
| + | first, BSimApp, is an interactive graphical user interface that allows for | ||
| + | simulation definitions to be displayed and run. For large scale simulations this approach is | ||
| + | not ideal and so the BSimBatch interface permits simulations to be run in a batch mode with | ||
| + | no user involvement. BSimBatch was extensively used to run simulations on the Blue Crystal | ||
| + | supercomputer at the university. | ||
| - | + | To specify a simulation, standard text based files are used to hold all parameter values and | |
| + | the environment configuration. Entries are grouped into the following main areas: | ||
| + | * General Simulation Parameters - Includes aspects such as time step, medium viscosity, potential function definitions, default size of bacteria/particles, bacterium forces and run lengths. | ||
| + | * Bacteria, Particles and Boundary Sets - Any number of objects can be defined in the simulated environment. Bacteria and particles can be created and placed individually, or there is the option of generating sets of randomly distributed groups within a specified area. Boundaries are defined by a start and end point and can be either solid or wrapping, where an offset vector is given to determine the new location of objects after they have crossed the boundary. | ||
| + | * Chemical Fields - Diffusion of chemicals in the environment is handled by 3 separate chemical fields representing the goal chemoattractant, short range co-ordination signal and long range recruitment chemoattractant. Each of these can be configured independently allowing for location, resolution, boundary type and diffusion co-efficient to be altered. There is also the option of having fixed gradients, minimising the mathematical calculations that need to be carried out. | ||
| + | * Visual Aids - Several visual aids have been provided to allow for easier analysis of videos. These include particle and bacteria tracers for individual objects or average positions, a simulation clock and scale. | ||
| + | * Simulation View - Includes the size of the simulation screen, zoom factors and the translation vector for the view. | ||
| + | * Batch Settings - To allow for running on headless computers, like the Blue Crystal supercomputer, it is possible to pre-define the simulation length, output path of data files, and the number of runs that need to be performed. | ||
=== User Interface === | === User Interface === | ||
| Line 37: | Line 59: | ||
The command line based batch interface is also known as BSimBatch. This provides a more advanced way to run large numbers of simulations and to handle the output files of statistics from these. Because the output from BSim is stochastic due to the way bacteria move, it is necessary to perform large numbers of simulation runs to gain statistically significant results. BSimBatch allows for this using the same simulation definition file as BSimApp but adding a few additional parameters such and number of runs and export directories. BSimBatch has also been designed to make it easy to run simulations on large computing clusters as jobs can be scheduled to run with no user intervention required. | The command line based batch interface is also known as BSimBatch. This provides a more advanced way to run large numbers of simulations and to handle the output files of statistics from these. Because the output from BSim is stochastic due to the way bacteria move, it is necessary to perform large numbers of simulation runs to gain statistically significant results. BSimBatch allows for this using the same simulation definition file as BSimApp but adding a few additional parameters such and number of runs and export directories. BSimBatch has also been designed to make it easy to run simulations on large computing clusters as jobs can be scheduled to run with no user intervention required. | ||
| + | |||
| + | === Defining Simulations === | ||
| + | |||
| + | Everything related to a simulation in BSim is contained within the parameter file. This text based file allows for environmental parameters (viscosity, bacteria speed, etc) to be set and for the simulated environmnet to be defined (locations of walls, initial placement of bacteria, etc). For a full list of the items that can be amnipulated in these files see the [[Team:BCCS-Bristol/Modeling-BSim_Parameter_File|Parameter File Definition]] page. | ||
=== Batch Simulations === | === Batch Simulations === | ||
| Line 59: | Line 85: | ||
* Scatter plots. | * Scatter plots. | ||
* Means. | * Means. | ||
| - | |||
| - | |||
| - | |||
<br> | <br> | ||
</div> | </div> | ||
Latest revision as of 17:18, 28 October 2008
Contents |
Stochastic Agent Based Simulation
Welcome to BSim
The stochastic agent based simulation framework we have developed is called BSim. This allows for differing configurations of particles and bacteria to easily be modelled in an interactive way. The framework has been developed from scratch in Java allowing it to be run on the majority of computer systems. In addition to an easy to use GUI, a command line based interface is also provided to permit the running of large scale simulations on remote computing clusters.
Framework Design
The BSim framework is a cross platform Java based programming API that has been developed from scratch to model the stochastic interactions of bacteria and particles in a fluid based environment. Taking this approach allowed for the greatest flexibility including features essential to our models. While this lead to an initially large effort in getting a working system completed, it later significantly increased the speed at which additional aspects could be incorporated. BSim has been developed using an object-oriented methodology and split into several highlevel modules shown in Figure 2.5. To give end-users the option of running the system on headless computers, e.g. via remote terminals, two alternative interfaces were created. The first, BSimApp, is an interactive graphical user interface that allows for simulation definitions to be displayed and run. For large scale simulations this approach is not ideal and so the BSimBatch interface permits simulations to be run in a batch mode with no user involvement. BSimBatch was extensively used to run simulations on the Blue Crystal supercomputer at the university.
To specify a simulation, standard text based files are used to hold all parameter values and the environment configuration. Entries are grouped into the following main areas:
- General Simulation Parameters - Includes aspects such as time step, medium viscosity, potential function definitions, default size of bacteria/particles, bacterium forces and run lengths.
- Bacteria, Particles and Boundary Sets - Any number of objects can be defined in the simulated environment. Bacteria and particles can be created and placed individually, or there is the option of generating sets of randomly distributed groups within a specified area. Boundaries are defined by a start and end point and can be either solid or wrapping, where an offset vector is given to determine the new location of objects after they have crossed the boundary.
- Chemical Fields - Diffusion of chemicals in the environment is handled by 3 separate chemical fields representing the goal chemoattractant, short range co-ordination signal and long range recruitment chemoattractant. Each of these can be configured independently allowing for location, resolution, boundary type and diffusion co-efficient to be altered. There is also the option of having fixed gradients, minimising the mathematical calculations that need to be carried out.
- Visual Aids - Several visual aids have been provided to allow for easier analysis of videos. These include particle and bacteria tracers for individual objects or average positions, a simulation clock and scale.
- Simulation View - Includes the size of the simulation screen, zoom factors and the translation vector for the view.
- Batch Settings - To allow for running on headless computers, like the Blue Crystal supercomputer, it is possible to pre-define the simulation length, output path of data files, and the number of runs that need to be performed.
User Interface
Two different user interface exist in BSim to serve the varying requirements when designing and running simulations. The first is an interactive graphical user interface (GUI) which provides easy to use controls for loading, running and exporting single simulations. The second is a command line based batch interface. This is provides functionality to allow for batches of simulations to be run in the background or on large cluster based computing systems, such as Blue Crystal.
BSimApp - Interactive Graphical User Interface (GUI)
The interactive graphical user interface to BSim is also know as BSimApp. It is a single user program that enables visual loading, running and exporting of simulations. It's main purpose is to allow for a single user to experiment with different parameters and simulation setups (e.g. number of bacteria, locations of objects, boundaries, etc) and to see any obvious problems that may arise before running the simulation hundreds of times to gain statistically significant results. It also provides a nice environment in which to debug any new simulation features that have been written, such as new bacteria types or simulation objects.
BSimBatch - Command Line Batch Interface
The command line based batch interface is also known as BSimBatch. This provides a more advanced way to run large numbers of simulations and to handle the output files of statistics from these. Because the output from BSim is stochastic due to the way bacteria move, it is necessary to perform large numbers of simulation runs to gain statistically significant results. BSimBatch allows for this using the same simulation definition file as BSimApp but adding a few additional parameters such and number of runs and export directories. BSimBatch has also been designed to make it easy to run simulations on large computing clusters as jobs can be scheduled to run with no user intervention required.
Defining Simulations
Everything related to a simulation in BSim is contained within the parameter file. This text based file allows for environmental parameters (viscosity, bacteria speed, etc) to be set and for the simulated environmnet to be defined (locations of walls, initial placement of bacteria, etc). For a full list of the items that can be amnipulated in these files see the Parameter File Definition page.
Batch Simulations
Batch simulations allow us to perform statistical analysis on our simulations and in particular look at the affect of altering one or many of the parameters. This will enable us to inform the wet lab team of the optimum conditions for their experiments.
Statistical Analysis
For every simulation statistical analysis will be performed on six aspects:
- The direction (angle) the particle has moved towards the chemoattractant.
- The distance (x axis) travelled by the particle.
- The mean direction (angle) of movement by all particles towards the chemoattractant.
- The mean distance (x axis) travelled by all particles.
- The mean direction (angle) of bacterial movement towards the chemoattractant
- The mean distance (x axis) bacteria travelled.
For each altered parameter 100 simulations (if feasible) will be run with each simulation running for 10 000 time steps containing 1-10 particles.
Methods of Statistical Analysis
Methods of Statistical analysis will include:
- T tests to show that results observed due to parameter changes are statistically significant.
- Histograms.
- Scatter plots.
- Means.
 "
"