Team:BrownTwo/Limiter/network
From 2008.igem.org
| Line 57: | Line 57: | ||
<p> | <p> | ||
| - | To | + | To solve this problem of interference with normal levels of transcription, we need a way to turn our repressor off when activation falls below a certain level, the threshold of healthy expression. |
</p> | </p> | ||
| - | |||
==The Bistable Switch== | ==The Bistable Switch== | ||
Revision as of 11:55, 29 October 2008
The Genetic Limiter
Our limiter circuit is an example of a complex device built from smaller parts and circuits in the mode of the synthetic biologist. To push the limits of synthetic biology beyond the conventional bacterial chassis, we have elected to build our device in Saccharomyces cerevisiae, an oft-studied model eukaryote that has heretofore been quite negelected in iGEM competition. Leading to the limiter
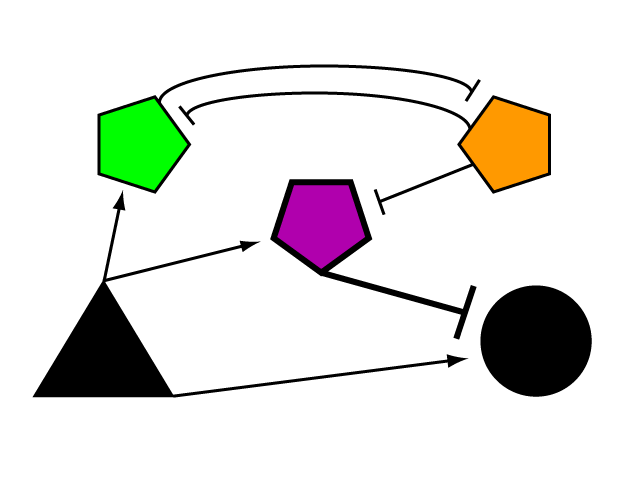
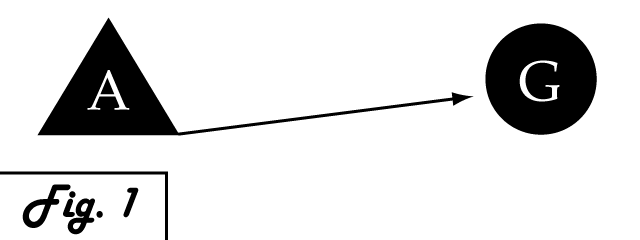
Consider a gene of interest (G) for which one inducing transcription factor (A) is known (Fig. 1). In the presence of unnaturally-heightened levels of inducer A, our gene will experience excessive activation. A logical method for addressing this problem would be to introduce an inhibitor for the gene of interest to attenuate its presence or activity. Traditional pharmaceutical interventions either block the activity of the protein encoded by gene G, causing interference on the level of protein interactions, or make changes upstream then can arrest expression of gene G. One concern that arises with the use of a chemical input is that, especially in the case of multi-cellular organisms, this often has diffuse, unspecified, and often undesirable effects. Drugs that are administered to biological systems in a therapeutic context particularly require an understanding of the intricate balance between efficacy and toxicity. An alternative may be to directly reduce the expression of gene G with an introduced transcriptional repressor. Although a genetically-based inhibitor is more difficult to implement, such a genetic tool would afford a great deal of specificity to the system, directly repressing only the gene of interest.
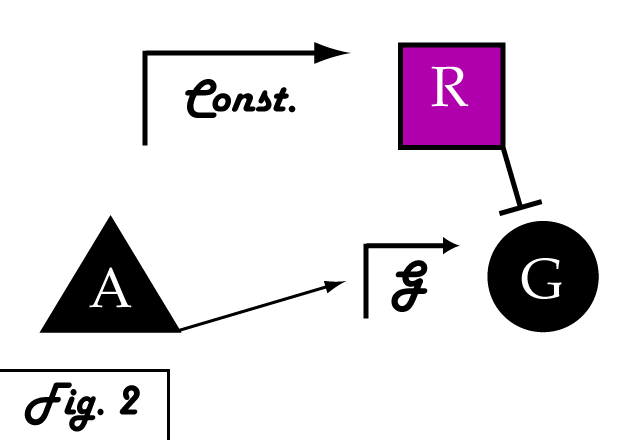
If we were to introduce a repressing transcription factor (R) driven by a constitutive promoter (Fig. 2), we would see a constant amount of repression of G, independent of its level of activation. We might face the issue that increased expression of inducer A would surpass the repressor's ability to oppose gene G's activation. Thus, instead of a constitutive promoter, we could ensure that the repressor for the gene of interest is under the control of a promoter that also takes the same inducer A as an input (Fig. 3). This would mean that the amount of repressor expressed in the system scales with the amount of inducer, eliminating the possibility of our repressor being swamped by incresing induction of G's transcription. However, both of these constructs have a critical flaw- they would always repress gene G, even when and where its expression is at a normal, healthy level. Repressing gene G completely could cause problems by eliminating the possibility of gene G functioning normally.
To solve this problem of interference with normal levels of transcription, we need a way to turn our repressor off when activation falls below a certain level, the threshold of healthy expression.
The Bistable SwitchA bistable switch is a simple circuit in which, depending upon the nature or quantity of the input, two stable outcomes are possible. In one case, would allow the system to alternate between two modes of activity in response to varying levels of input. Dr. James Collins from Boston University developed a bistable switch capable of switching between two genetic outputs based upon the inputs to the system. Driven by the desire to create a synthetic gene network that could act similarly to endogenous regulatory pathways, we developed a design for a system that had the ability to regulate We began by developing a layout for
Diagram our network and explain why we laid it out this way.
|
 "
"