Team:Hawaii/Milestone
From 2008.igem.org
(Difference between revisions)
Normanwang (Talk | contribs) (update) |
|||
| (26 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
| + | {{Team:Hawaii/Header}}[[Image:20080612-work breakdown structure.jpg|thumb|300px|right]] | ||
== Immediate == | == Immediate == | ||
| + | <onlyinclude> | ||
| + | * Culturing Protocol Optimization: CO2 is bottle neck, need incubator retrofitted (KS) | ||
| + | * Express GFP constructs in ''Synechocystis'' | ||
| + | </onlyinclude> | ||
| + | == Near Term == | ||
| - | * | + | == Completed Milestones == |
| - | * | + | * 2008.05.xx figure out OD Reading operation: Beckman 640U spectrophotometer (AB/NW) |
| - | + | * [[Team:Hawaii/Notebook/2008-05-22|2008.05.24]] Create seed stocks for test transformation (NW/MR) | |
| - | + | * [[Team:Hawaii/Meeting/2008-05-22|2008.05.22]] Describe, present [http://partsregistry.org/Part_Types part types]. 1. Norman: biobrick assembly, parts, 2. Margaret: devices, 3. Krystle: chassis & plasmids, 4. Grace: other | |
| - | + | * 2008.06.01 Plasmid Transformation Plan/Protocol (GK/MR) | |
| - | + | * 2008.06.02 Create competent DH5a E. coli stocks (GK/MR/NW) | |
| - | + | * 2008.06.03-2008.06.04 Plasmid Transformation, document results (GK/NW) | |
| - | + | * 2008.06.04 GK and KS are now autoclave trained | |
| - | + | * 2008.06.10 Fluorometer found (in Dr. Su's lab), 470nm blue LED lights purchased for constructing our own light table. | |
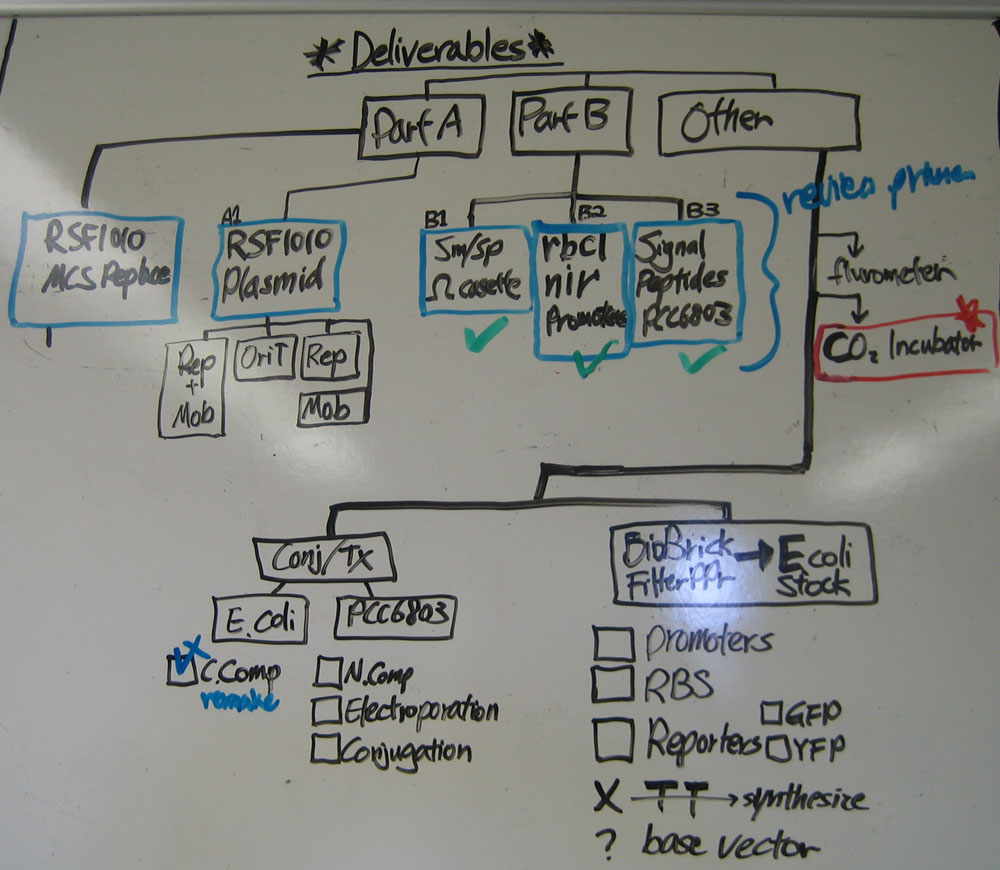
| - | [[Team:Hawaii/Meeting/2008- | + | * 2008.06.11 First draft Work Breakdown Structure diagram drawn on whiteboard, detailing all deliverables. (please keep it updated) |
| - | + | * 2008.06.11 Does RSF1010 replicate autonomously in PCC6803? YES, only when it goes in cell as circle. linear DNA need recombination. Determined we need to test all three methods to find one that works: 1. Chemical 2. Electroporation 3. Conjugation (MR) | |
| - | + | * 2008.06.12 Final draft of proposal due | |
| - | + | * 2008.06.23 Transform PCC6803 w/ pRL1383a | |
| - | + | * 2008.06.24 First batch of primers ordered | |
| - | + | * 2008.06.24 Biobrick parts list completed | |
| - | + | * 2008.06.24 RP1 and RP4 oriT are the same | |
| - | + | * 2008.08.13 Replaced pRL1383a MCS with BioBrick MCS (GK) | |
| - | + | {{Team:Hawaii/Footer}} | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
Latest revision as of 17:35, 15 August 2008
| Projects | Events | Resources | ||
|---|---|---|---|---|
| Sponsors | Experiments | Milestones | Protocols | |
| Notebook (t) | Meetings (t) |
Immediate
- Culturing Protocol Optimization: CO2 is bottle neck, need incubator retrofitted (KS)
- Express GFP constructs in Synechocystis
Near Term
Completed Milestones
- 2008.05.xx figure out OD Reading operation: Beckman 640U spectrophotometer (AB/NW)
- 2008.05.24 Create seed stocks for test transformation (NW/MR)
- 2008.05.22 Describe, present [http://partsregistry.org/Part_Types part types]. 1. Norman: biobrick assembly, parts, 2. Margaret: devices, 3. Krystle: chassis & plasmids, 4. Grace: other
- 2008.06.01 Plasmid Transformation Plan/Protocol (GK/MR)
- 2008.06.02 Create competent DH5a E. coli stocks (GK/MR/NW)
- 2008.06.03-2008.06.04 Plasmid Transformation, document results (GK/NW)
- 2008.06.04 GK and KS are now autoclave trained
- 2008.06.10 Fluorometer found (in Dr. Su's lab), 470nm blue LED lights purchased for constructing our own light table.
- 2008.06.11 First draft Work Breakdown Structure diagram drawn on whiteboard, detailing all deliverables. (please keep it updated)
- 2008.06.11 Does RSF1010 replicate autonomously in PCC6803? YES, only when it goes in cell as circle. linear DNA need recombination. Determined we need to test all three methods to find one that works: 1. Chemical 2. Electroporation 3. Conjugation (MR)
- 2008.06.12 Final draft of proposal due
- 2008.06.23 Transform PCC6803 w/ pRL1383a
- 2008.06.24 First batch of primers ordered
- 2008.06.24 Biobrick parts list completed
- 2008.06.24 RP1 and RP4 oriT are the same
- 2008.08.13 Replaced pRL1383a MCS with BioBrick MCS (GK)
[http://manoa.hawaii.edu/  ][http://manoa.hawaii.edu/ovcrge/
][http://manoa.hawaii.edu/ovcrge/  ][http://www.ctahr.hawaii.edu
][http://www.ctahr.hawaii.edu  ]
]
 "
"