Team:Heidelberg/Notebook/Killing I/Notebook/week3
From 2008.igem.org
| Line 165: | Line 165: | ||
** Analytical digestion of cI "green", cI "black" and T9002 | ** Analytical digestion of cI "green", cI "black" and T9002 | ||
| - | *Gel | + | *Gel |
[[Image:Hd-phage-08-08-19-digestion.jpg]] | [[Image:Hd-phage-08-08-19-digestion.jpg]] | ||
#* Lane 0: DNA ladder mix | #* Lane 0: DNA ladder mix | ||
| Line 197: | Line 197: | ||
[[Image:Hd-phage-wachstumskurve_errorbars_small.jpg]] | [[Image:Hd-phage-wachstumskurve_errorbars_small.jpg]] | ||
**the values are the average of all four cultures | **the values are the average of all four cultures | ||
| - | + | ||
| Line 223: | Line 223: | ||
===project planning=== | ===project planning=== | ||
| - | Guys, we received this email by another professor in the states. sounds good. we should get this helper plasmids in the end :) | + | Guys, we received this email by another professor in the states. sounds good. we should get this helper plasmids in the end :) |
| - | + | ||
I have sent pUB307, RSF1010, pED350, pED361, which are described in our two MGG papers. Also two clones I made of the oriT from RP1; pED369 and pED374. There are 10microl of each. The DNAs are old (1983!) and so I would transform and make | I have sent pUB307, RSF1010, pED350, pED361, which are described in our two MGG papers. Also two clones I made of the oriT from RP1; pED369 and pED374. There are 10microl of each. The DNAs are old (1983!) and so I would transform and make | ||
fresh plasmid preps. | fresh plasmid preps. | ||
| - | + | ||
pED350 is oriT+ and Mob+, so it is efficiently mobilised by RPI or its derivative pUB307. pED361 is oriT+ only. It cannot be mobilised by pUB307 as it lacks mob functions. If RSF1010 is also in the cell pKD361 is very efficiently mobilised by pUB307. | pED350 is oriT+ and Mob+, so it is efficiently mobilised by RPI or its derivative pUB307. pED361 is oriT+ only. It cannot be mobilised by pUB307 as it lacks mob functions. If RSF1010 is also in the cell pKD361 is very efficiently mobilised by pUB307. | ||
| - | + | ||
pED369 and pED374 are two subclones I made of the RP1 oriT into pED825 (Amp resistant described in MGG papers). pED374 contains a single 690 bp HaeII fragment containing the | pED369 and pED374 are two subclones I made of the RP1 oriT into pED825 (Amp resistant described in MGG papers). pED374 contains a single 690 bp HaeII fragment containing the | ||
RP1 oriT. PED369 contains the 690 bp oriT fragment plus additional HaeII fragments. I sent both just in case the pED374 DNA does not check out. These are efficiently | RP1 oriT. PED369 contains the 690 bp oriT fragment plus additional HaeII fragments. I sent both just in case the pED374 DNA does not check out. These are efficiently | ||
mobilised by pUB307. | mobilised by pUB307. | ||
| - | + | ||
I would think cloning this 690 bp fragment into your lambda vector would be the simplest | I would think cloning this 690 bp fragment into your lambda vector would be the simplest | ||
solution; its small and will mobilise in the presence of just pUB307. | solution; its small and will mobilise in the presence of just pUB307. | ||
| - | + | ||
Keith | Keith | ||
| - | + | ||
| Line 295: | Line 295: | ||
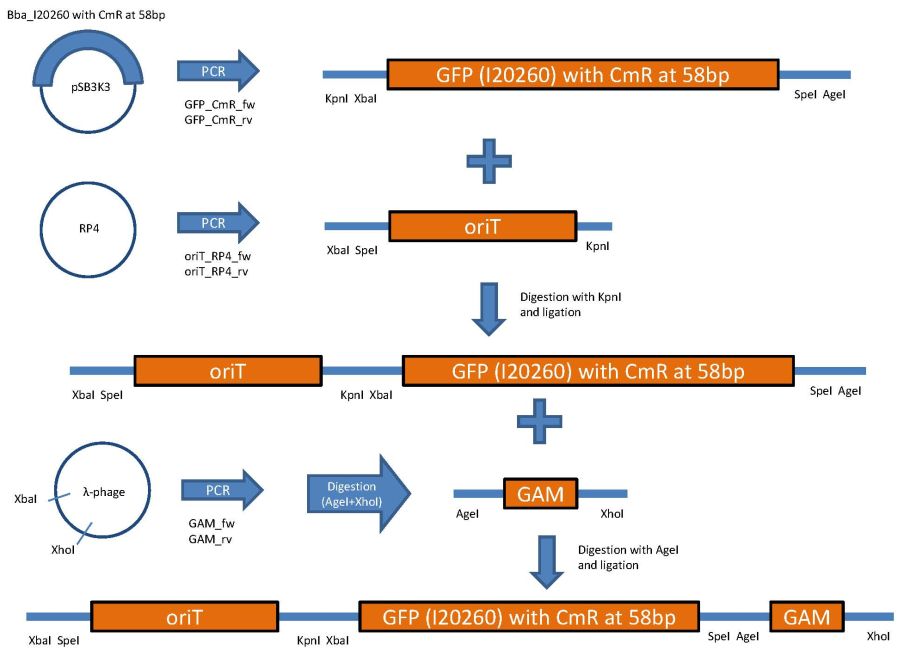
[[Image:Hd-phage-Klonierungsstrategie_Page_3.jpg]] | [[Image:Hd-phage-Klonierungsstrategie_Page_3.jpg]] | ||
<br><br><br> | <br><br><br> | ||
| - | + | ||
| - | + | ||
<br><br><br> | <br><br><br> | ||
Used primer sequences: | Used primer sequences: | ||
| - | + | ||
oriT_RP4_fw (Tm=62°C): | oriT_RP4_fw (Tm=62°C): | ||
| Line 306: | Line 305: | ||
CTCGTTTCTAGAACTAGTgacaggctcatgccggccgc | CTCGTTTCTAGAACTAGTgacaggctcatgccggccgc | ||
| - | + | ||
oriT_RP4_rv: (Tm=58°C) | oriT_RP4_rv: (Tm=58°C) | ||
TATTCGGGTACCgtcccctcagttcagtaatttcctgc | TATTCGGGTACCgtcccctcagttcagtaatttcctgc | ||
| - | + | ||
GFP_CmR_fw: (Tm=56°C) | GFP_CmR_fw: (Tm=56°C) | ||
CTCGTTGGTACCTCTAGAtttacagctagctcagtcctagg | CTCGTTGGTACCTCTAGAtttacagctagctcagtcctagg | ||
| - | + | ||
GFP_CmR_rv: (Tm=55°C) | GFP_CmR_rv: (Tm=55°C) | ||
TATTCGACCGGTACTAGTtataaacgcagaaaggcccacc | TATTCGACCGGTACTAGTtataaacgcagaaaggcccacc | ||
| - | + | ||
GAM_fw: (Tm=55°C) | GAM_fw: (Tm=55°C) | ||
AGTGCTTTAGCGTTAACTTCCG | AGTGCTTTAGCGTTAACTTCCG | ||
| - | + | ||
GAM_rv: (Tm=53°C) | GAM_rv: (Tm=53°C) | ||
GGTTTTACCGCATACCAATAACG | GGTTTTACCGCATACCAATAACG | ||
| - | + | ||
CmR_fw: (Tm=54°C) | CmR_fw: (Tm=54°C) | ||
gctaaaATGgagaaaaaaatcactgg | gctaaaATGgagaaaaaaatcactgg | ||
| - | + | ||
CmR_rv: (Tm=58°C) | CmR_rv: (Tm=58°C) | ||
AGGTTCTCCTTTATTAGCCGGATCCTCTAGATTACGCC | AGGTTCTCCTTTATTAGCCGGATCCTCTAGATTACGCC | ||
| - | + | ||
Revision as of 22:05, 25 October 2008


Contents |
Monday, 08/18/08
chloramphenicol resistance cassette
- Maxiprep of P1000, P1004, B0014, B0015
- P1000 1672 ng/µl 1,92
- P1004 1153 ng/µl 1,92
- B0014 2040,4 ng/µl 1,92
- B0015 1053,6 ng/µl 1,92
- Analytical digestions
- lambda DNA with XbaI and XhoI
2µl DNA 5µl NEB2 5µl BSA 1µl XbaI 1µl XhoI 26µl water
- Analytical digestion of P1000
1µl DNA 2µl NEB3 2µl NcoI 15µl water
- Analytical digestion of P1004
1µl DNA 2µl NEB4 1,5µl DraI 15,5µl water
- Analytical digestion of B0014 / B0015
1µl DNA 2µl NEB4 2µl BSA 2,5µl SfcI 12,5µl water
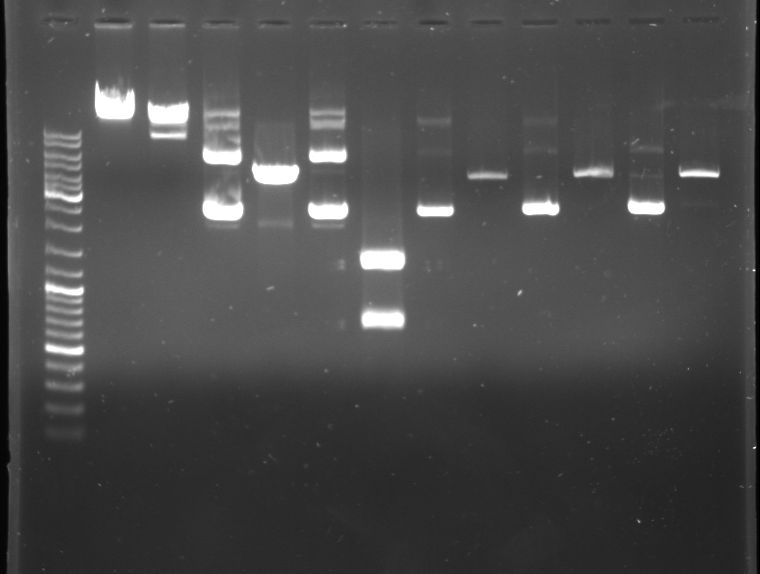
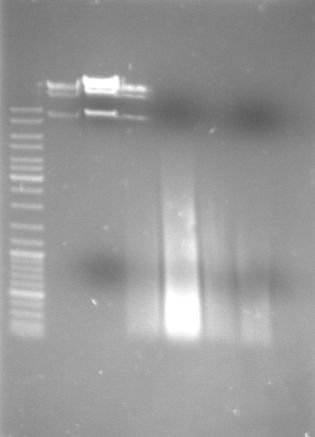
- Analytical digestion of cI "green", cI "black" and T9002
- Gel
- Lane 0: DNA ladder mix
- Lane 1: lambda DNA
- Lane 2: lambda DNA (XbaI, XhoI --> large frament ~ 40kb, small fragment ~ 9kb)
- Lane 3: P1000
- Lane 4: P1000 (NcoI-->1473,2374)
- Lane 5: P1004
- Lane 6: P1004 (DraI-->19,339,692,1067,1360)
- Lane 7: cI green
- Lane 8: cI green (NdeI - expected fragments?)
- Lane 9: cI black
- Lane 10: cI black (NdeI)
- Lane 11: T9002
- Lane 12: T9002 (NdeI - expected fragments?)
lamda phage
Infectiontest
- inoculation of 20ml medium with over night culture (MG1655)
- after 1h plaque added --> 37C for 2h + inoculation of ZMBH phage (100ql 10^-2 in 20ml MG1655)
- incubation at 42C for 2 more hours
- sterilefiltration + mix with growing bacteria (MG1655 and MG1655+cI)
- incubate in soft agar at 37C for 2h, than at 42C for another 4h (altogether 24 plates)
- phage DNA was also isolated from filtrate
- results:
- same amount of plaques on plates with MG1655 as with MG1655+cI
- nearly no plaques on plates with own phage --> very low phage concentration
- --> cI does not work!!!!
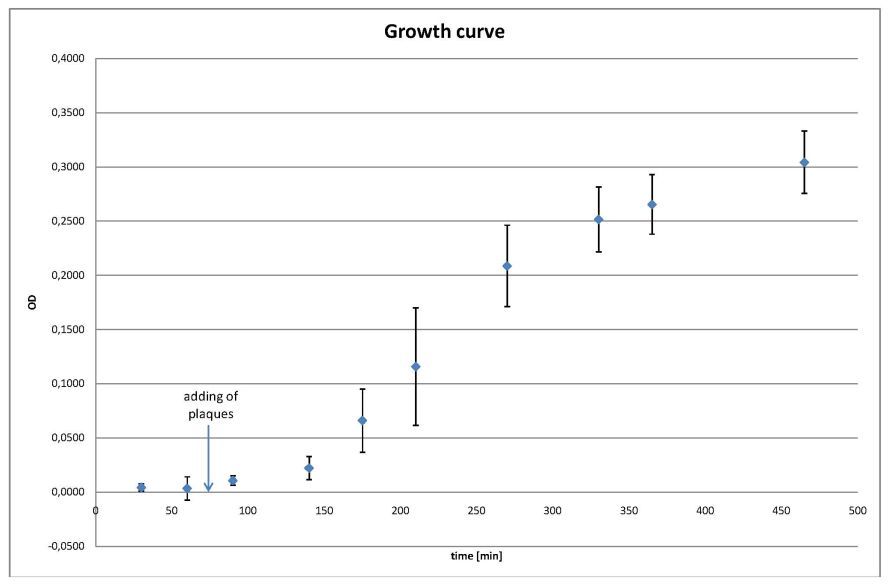
- growing curve
- the values are the average of all four cultures
Tuesday, 08/19/08
lambda phage
- Digestion of lambda DNA with XbaI and XhoI
- very nice separation of the small fragment from the two large fragments (one band) picture of the gel was not saved but it looked very nice
- cutting out of the small and large fragment --> gel purification kit
- small fragment: 30,3 ng/µl; 1,86
- large fragment1: 19,5 ng/µl; 2,11
- large fragment2: 61,5ng/µl; 1,90
GFP
- inoculation of three overnight cultures from the two I20260 glycerol stocks
chloramphenicol resistance cassette
- Transformation of P1000, P1004 (both chloramphenicol resistances) and CI
- no success
project planning
Guys, we received this email by another professor in the states. sounds good. we should get this helper plasmids in the end :)
I have sent pUB307, RSF1010, pED350, pED361, which are described in our two MGG papers. Also two clones I made of the oriT from RP1; pED369 and pED374. There are 10microl of each. The DNAs are old (1983!) and so I would transform and make fresh plasmid preps.
pED350 is oriT+ and Mob+, so it is efficiently mobilised by RPI or its derivative pUB307. pED361 is oriT+ only. It cannot be mobilised by pUB307 as it lacks mob functions. If RSF1010 is also in the cell pKD361 is very efficiently mobilised by pUB307.
pED369 and pED374 are two subclones I made of the RP1 oriT into pED825 (Amp resistant described in MGG papers). pED374 contains a single 690 bp HaeII fragment containing the RP1 oriT. PED369 contains the 690 bp oriT fragment plus additional HaeII fragments. I sent both just in case the pED374 DNA does not check out. These are efficiently mobilised by pUB307.
I would think cloning this 690 bp fragment into your lambda vector would be the simplest solution; its small and will mobilise in the presence of just pUB307.
Keith
Wednesday, 08/20/08
GFP
- Miniprep of reference promotor I20260
- Concentrations:
- Epi1 9,8 ng/ul; 2,52
- Epi2 15,7 ng/ul; 2,28
- Epi3 19,7 ng/ul; 1,93
- Concentrations:
- Digestion of I20260
10 µl DNA (from Epi3) 2ul SmlI 12µl NEB4 2ul BSA 10x 4 µl water
- Digestion of the XbaI-XhoI fragment with AgeI
18 µl DNA (from gel purification kit) 5 µl AgeI 5 µl NEB4 22 µl water
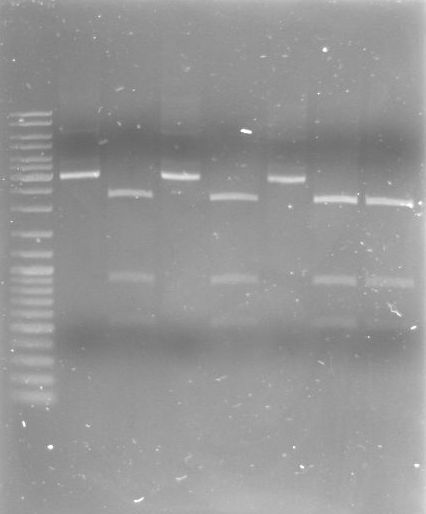
- Gel
- Lane 0: DNA ladder mix
- Lane 1: I20260 (SmlI-->251bp, 373bp, 843bp, 918bp, 1284bp)
- Lane 2: XbaI-XhoI fragment (AgeI-->1.7kb, 7.3kb)
- Lane 3: large lambda DNA fragments 3µl (15kb, 24.5kb)
- Results:
- I20260 seems not to be correct, inoculate overnight cultures from the glycerol stocks again
- digestion of the XbaI-XhoI fragment with AgeI did not work good, only the 7.3kb fragment can be seen, reasons: wrong buffer concentration, too much enzyme (glycerol) in the reaction
- large lambda DNA fragment is looking good
lambda phage
- concentrations of lambda-DNA extractions
- from Fermentas Phage - plaque 1: 7.5 ng/ul; 1.86
- from Fermentas Phage - plaque 2: 61.5 ng/ul 1.90
- from ZMBH phage 19,7 ng/ul 2,11
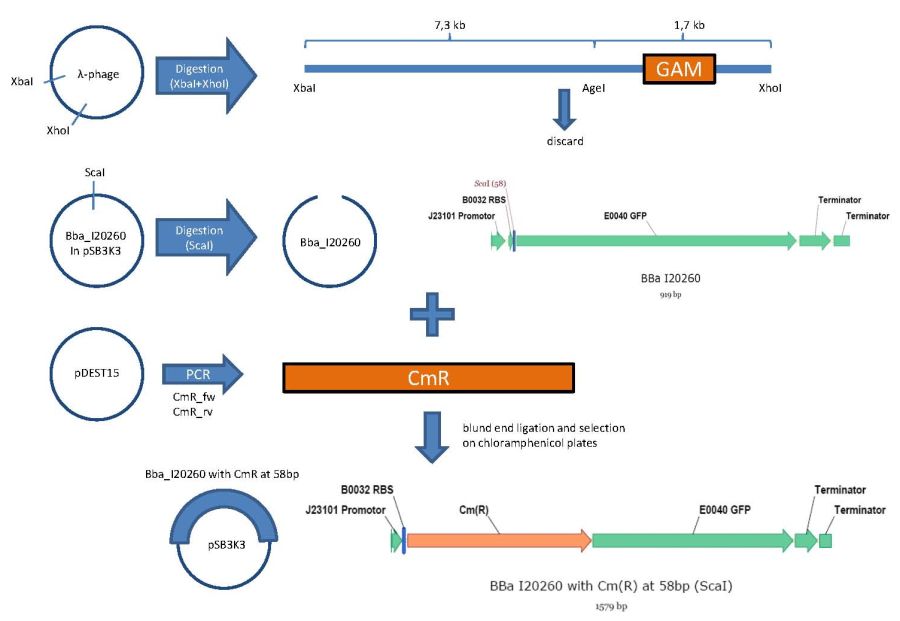
overview of the first phage cloning strategy
Used primer sequences:
oriT_RP4_fw (Tm=62°C):
CTCGTTTCTAGAACTAGTgacaggctcatgccggccgc
oriT_RP4_rv: (Tm=58°C)
TATTCGGGTACCgtcccctcagttcagtaatttcctgc
GFP_CmR_fw: (Tm=56°C)
CTCGTTGGTACCTCTAGAtttacagctagctcagtcctagg
GFP_CmR_rv: (Tm=55°C)
TATTCGACCGGTACTAGTtataaacgcagaaaggcccacc
GAM_fw: (Tm=55°C)
AGTGCTTTAGCGTTAACTTCCG
GAM_rv: (Tm=53°C)
GGTTTTACCGCATACCAATAACG
CmR_fw: (Tm=54°C)
gctaaaATGgagaaaaaaatcactgg
CmR_rv: (Tm=58°C)
AGGTTCTCCTTTATTAGCCGGATCCTCTAGATTACGCC
Thursday, 08/21/08
lambda phae
- Preparative digestion of lambda-DNA from Fermentas
10µl DNA = 3 ug 2µl XhoI 2µl XbaI 5µl NEB 2 5µl BSA 33µl H20
- lambda DNA "2" and "ZMBH" from phage extraction kit
36 µl DNA 2 µl XhoI 2µl XbaI 5µl NEB 2 5 µl BSA
- all digestions 3h, 400rpm, 37°
- Gel
- lane 0: DNA ladder mix
- lane 2: lambda DNA (fermentas) (XbaI, XhoI --> ~40 kb, ~9 kb)
- lane 4: lambda DNA from extraction "2" (XbaI, XhoI --> ~40 kb, ~9 kb)
- lane 6: lambda DNA from extraction "ZMBH phage" (XbaI, XhoI --> ~40 kb, ~9 kb)
- lane 2, 4 and 6 overflowed
- cutting out of the XbaI-XhoI fragment from lane 2
- the large fragment wasn't cutted out because the gel was exposed to UV light over 30 seconds
- gel extraction kit
- DNA eluted in 20µl
- Digestion of the XbaI-XhoI fragment with AgeI
20µl DNA (from gel purification kit) 2µl AgeI 5µl NEB4 23µl water
- 16°C over night
- Preparative digestion of lambda-DNA from Fermentas
10µl DNA = 3 µg 2µl XhoI 2µl XbaI 5µl NEB 2 5µl BSA 33µl water
- 16°C over night
GFP
- Miniprep from overnight cultures of I20260 glycerol stocks
- eluted in 50 µl
- Concentrations:
- Miniprep 1: 19.8 ng/µl
- Miniprep 2: 17.4 ng/µl
- Miniprep 3: 19.0 ng/µl
- Miniprep 4: 16.4 ng/µl
- Analytical digestion of the four I20260 Minipreps
- 20µl DNA
- 5µl NEB 4
- 1.5µl DraI
- 23.5µl water
- Gel
- lane 0: DNA ladder
- lane 1: I20260 1
- lane 2: I20260 1 (DraI-->19bp, 535bp, 886bp, 2229bp)
- lane 3: I20260 2
- lane 4: I20260 2 (DraI)
- lane 5: I20260 3
- lane 6: I20260 3 (DraI)
- lane 7: I20260 4 (DraI)
- Results
- it seems that we have I20260, inoculation of an overnight culture from I2020 No. 3 for Maxiprep
- Results
- Transformation of I20260 (Kan) (from the promotor measurement kit sheet as well as from the registry) and J01101 (Amp)
- eluted in 10µl water
- transformed into TOP10 and MG1655 (each 4µl DNA)
cI
- ordered J01101 (cI) from the iGEM headquater, they will already send it today
Friday, 08/22/08
GFP
- Maxiprep of reference promotor I20260
- concentrations: 200 ng/µl
lambda phage
- Gel: Digestions of lambda-DNA (fermentas), and the small XbaI-XhoI fragments
- lane 1: DNA ladder mix
- lane 3: lambda-DNA (XbaI, XhoI; ~ 40kb, ~ 9kb)
- lane 5: small lambda-fragment assayI (AgeI, ~1,7kb, ~7,3kb)
- lane 7: small lambda-fragment assayII (AgeI, ~1,7kb, ~7,3kb)
- Extraction of XbaI-XhoI fragments (evaluation of samples with nanodrop)
- small fragment 1: 15,9 ng/µl; 2,41
- small fragment 2: 6,6 ng/µl; 2,02
- large fragment 1: 14,1 ng/µl; 2,75
- large fragment 1: 10,8 ng/µl; 2,86
- large fragment 1: 14,4 ng/µl; 1,99
- large fragment 1: 16,8 ng/µl; 2,14
 "
"