Team:Heidelberg/Notebook/Killing I/Notebook/week7
From 2008.igem.org
Revision as of 11:38, 29 October 2008 by Maximilian.hoerner (Talk | contribs)


| << Week 6 | Overview | Week 8 >> |
|---|
Week 7
Contents |
Monday, 09/15/08
conjugation test
- inoculation of liquid cultures for conjugation tests (cotransformation of our insert in pBlue + pUB307 worked)
- conjugation test with pBlue::lambda, pUB307 and zeocin, each 1,5 ml of liquid culture, mixed and incubated for 1h, plated on Kan,Cm,Zeo,
controls: pBlue::lambda, pUB307 plated on Kan,Cm,Zeo and Zeocin plated on Kan,Cm,Zeo
phage cloning strategy one
- gel for cutting out oriT and rest insert (after digestion of complete insert (NotI/XhoI) with KpnI)
- --> no bands visible!!!
- digestion of lambda DNA (XbaI/XhoI) --> cutting out big fragment
- digestion of insert in pBluescript again with XhoI/NotI/KpnI --> cut out oriT and rest insert
- digest oriT with XbaI
- testdigestion of pUB307 with different restriction enzymes to make sure it has the right size befor we possibly sequence it
Tuesday, 09/16/08
conjugation test
- conjugation test did not work: no colonies on plates, fortunately also no colonies on control plates
- results at the end of the day: on one plate there was a lawn of bacteria - to decide wether it were the right and only very slow grown or it was a contamination, we picked colonies and inoculated them in LB with amp+cm+zeo and kan+zeo, repsectively.
phage cloning strategy one
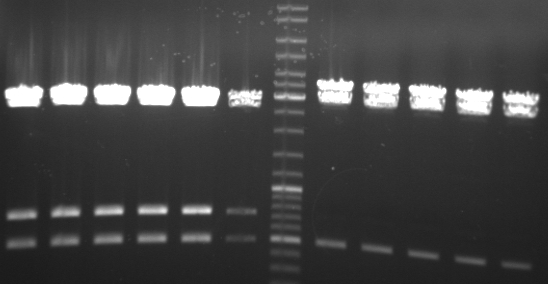
- gel result of testdigestion of pUB307:
- lane 0: DNA ladder mix
- lane 1: ScaI
- lane 2: PstI
- lane 3: XhoI
- lane 4: AgeI
- lane 5: BglI
- lane 6: NdeI
- lane 7: XbaI
- lane 8: XmaI
- lane 9: SfcI
- lane 10: EcoRI
- lane 11: SpeI
- lane 12: HindIII
- --> our plasmid seems to be the the one we have the sequence of (RP1 with deletion of tn1 from ncbi)
- literature on pUB307 and its sequence:
- pUB307 is a derivative of RP1, it's origin is a spontaneous mutation of RP1, which lost its Tn1 transposon containing an amp resistance
- Target Choice and Orientation Preference of the Insertion Sequence IS903 - http://jb.asm.org/cgi/content/abstract/180/12/3039
- digestion of pBlue+insert with XhoI/NotI/KpnI
- cut out oriT and rest insert
- ligation into lambda over night (16°C)
- nanodrop of lambda fragment gel extracted with EB for dilution
- 38,8 ng/l; 1,86
Wednesday, 09/15/09
conjugation test
- conjugation test with Zeocin and pUB307,pBlue::lambda
- 1,5ml of each, mix, incubate for 1h
- 1,5ml of each, fill up to 30ml with LB, incubate for 1h
- plate both with Amp, Cm, Zeo, incubate for 16-24 h (bacteria grow very slow, maybe because of the three antibiotics?)
- why dilute with LB: the problem is, if we put Zeo-Bacteria and pUB+pBlue bacteria together which were grown in media with the coresponding antibiotics, than the conjugation medium contains amp, cm and zeo, but the bacteria do not contain the resistance to all of them before conjugation. so the question is, if we kill many of our bacteria before they can even start to conjugate. Therefore we put LB to the conjugation medium to dilute the antibiotics.)
Thursday, 09/16/08
- new daily plan for the next days for mutagenesis
- new digestion, ligation and transformation of our insert in lambda
- conjugation test from 15.09.: the picked colonies did grow on amp,cm,zeo and on kan,zeo - so the first test worked
- conjugation test from 16.09. there were only colonies on the plated with the undiluted conjugation media. So it seems to be that the antibiotics do not influence conjugation - on the contrary, diluted with LB conjugation did not work
Friday, 09/17/08
phage cloning strategy one
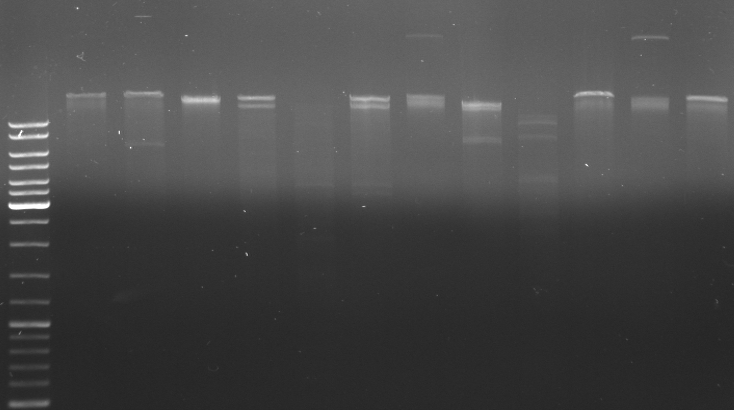
- new gel for cutting out our fragments: new approach: digest pBlue+insert with XbaI/KpnI (6 fragments) and cut out the fragment with 479bp (oriT). With this strategy we directly get the oriT with the correct ends. Next, we digested pBlue+insert with XhoI/KpnI (in this case also with NotI since we already did it yesterday) and extract the big fragment with the correct ends. With this approach we don't have to digest after the gel extraction again and therefore hope for better yields.
- lane1-6: digested with XbaI/KpnI --> cut out 479bp (oriT)
- lane7: DNA ladder mix
- lane8-12: digested with XhoI/KpnI/NotI --> cut out 3363bp (rest insert)
| << Week 6 | Overview | Week 8 >> |
|---|
 "
"