Team:Heidelberg/Notebook/Sensing Group/Cloning
From 2008.igem.org
(→Sensing - Cloning strategy) |
(→Sensing - Cloning strategy) |
||
| Line 124: | Line 124: | ||
==Sensing - Cloning strategy== | ==Sensing - Cloning strategy== | ||
| - | The core part of the sensing project is the construction of the LuxQ-Tar chimeric receptor, which enables the ''E. coli'' killer bacteria to chemotactically respond to a AI-2 gradient and detect prey cells. The quorum-sensing system | + | The core part of the sensing project is the construction of the LuxQ-Tar chimeric receptor, which enables the ''E. coli'' killer bacteria to chemotactically respond to a AI-2 gradient and detect prey cells. The quorum-sensing system will be amplified from the ''V. harveyi'' genome while the Tar receptor will be from ''E. coli''. On the one hand LuxS needs to be cloned and transformed into one cell type, to make them produce and secrete AI-2. On the other hand the periplasmic ligand binding domain will be fused to the cytoplasmic domain of Tar and cloned on one plasmid together with LuxP which is necessary for AI-2 binding. Generally, needed restriction sites for cloning are introduced via the PCR primer. |
''In silico'' cloning was performed in [http://serialbasics.free.fr/Serial_Cloner.html SerialCloner], Vector maps were designed with PlasMapper [1]. | ''In silico'' cloning was performed in [http://serialbasics.free.fr/Serial_Cloner.html SerialCloner], Vector maps were designed with PlasMapper [1]. | ||
| Line 132: | Line 132: | ||
=== LuxS === | === LuxS === | ||
| - | LuxS | + | LuxS will be amplified from the ''V. harveyi'' genome with primers LuxSa/LuxSb. Subsequently the product will be cloned into the pTr99alpha plasmid at the NcoI and BamHI sites and transformed into DH5a competent cells. |
=== LuxQ === | === LuxQ === | ||
| Line 142: | Line 142: | ||
=== Fusion receptor === | === Fusion receptor === | ||
| - | A detailed description of the Tar receptor and LuxQ quorum-sensing receptor can be found in the [[Team:Heidelberg/Project/Sensing | project description]]. Since it is believed that the linker region (TM2) is critically for signal transduction two different receptor constructs | + | A detailed description of the Tar receptor and LuxQ quorum-sensing receptor can be found in the [[Team:Heidelberg/Project/Sensing | project description]]. Since it is believed that the linker region (TM2) is critically for signal transduction two different receptor constructs are designed, referred to as Fusion-1 and Fusion-2. Fusion-1 contains the cytoplasmic domain of Tar and TM2 to the N-terminal of LuxQ (TM2, periplasmic domain, TM1). Fusion-2 contains the cytoplasmic domain and TM2 of Tar and the periplasmic domain to the N-terminal end of LuxQ (periplasmic domain, TM1). |
| - | The receptor chimeras | + | The receptor chimeras will be built in two sequential PCR reactions. In a first PCR the two single fragments containing parts of the coding sequence of LuxQ or Tar will be generated. The reverse primer for LuxQ and the forward primer for Tar have complementary ends, allowing annealing during the second PCR reaction. In addition to the amplified sequences from reaction 1 the forward primer for LuxQ and the reverse primer for Tar will be added to the reaction mix in order to obtain the chimeric receptor sequence. |
=== References === | === References === | ||
[1] Dong, X.; Stothard, P.; Forsythe, I. J. & Wishart, D. S., '''PlasMapper: a web server for drawing and auto-annotating plasmid maps''', Nucleic Acids Res, Vol. 32, pp. W660-W664, 2004 | [1] Dong, X.; Stothard, P.; Forsythe, I. J. & Wishart, D. S., '''PlasMapper: a web server for drawing and auto-annotating plasmid maps''', Nucleic Acids Res, Vol. 32, pp. W660-W664, 2004 | ||
Revision as of 21:48, 26 October 2008


Contents |
Sensing - Cloning strategy
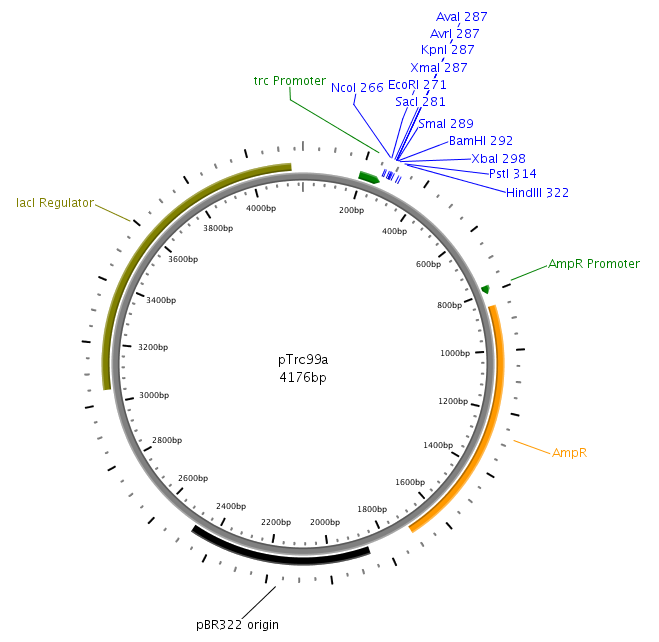
The core part of the sensing project is the construction of the LuxQ-Tar chimeric receptor, which enables the E. coli killer bacteria to chemotactically respond to a AI-2 gradient and detect prey cells. The quorum-sensing system will be amplified from the V. harveyi genome while the Tar receptor will be from E. coli. On the one hand LuxS needs to be cloned and transformed into one cell type, to make them produce and secrete AI-2. On the other hand the periplasmic ligand binding domain will be fused to the cytoplasmic domain of Tar and cloned on one plasmid together with LuxP which is necessary for AI-2 binding. Generally, needed restriction sites for cloning are introduced via the PCR primer. In silico cloning was performed in [http://serialbasics.free.fr/Serial_Cloner.html SerialCloner], Vector maps were designed with PlasMapper [1].
LuxS
LuxS will be amplified from the V. harveyi genome with primers LuxSa/LuxSb. Subsequently the product will be cloned into the pTr99alpha plasmid at the NcoI and BamHI sites and transformed into DH5a competent cells.
LuxQ
LuxQ will be amplied with primers LuxQa/LuxQc from the V. harveyi genome. The fragment will then be cloned into pTrc99alpha plasmid at the NcoI and BamHI site. Subsequently this construct will be used as template for constructing the Fusion receptors.
LuxP
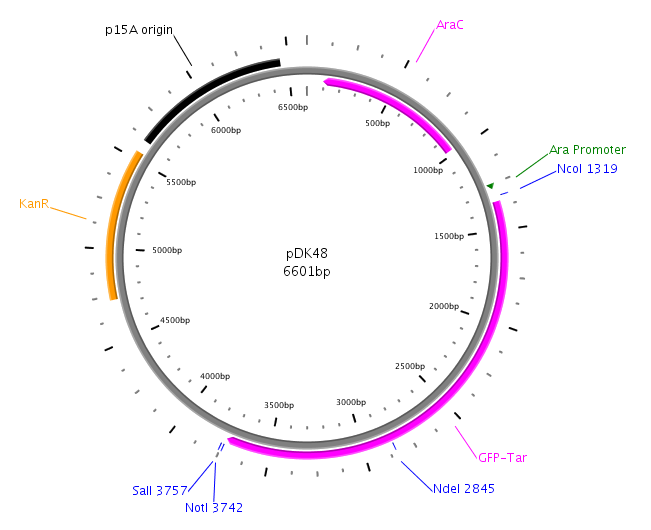
LuxP will be amplied with primers LuxPc/LuxPd from the V. harveyi genome. Then the product will be cloned into native pDK48. After the construction of the LuxQ-Tar Fusion gene in pDK48 plasmid, LuxP will also be introduced in the pDK48 plasmid at SalI and NotI sites. The whole construct of pDK48 will be transformed into our killer cell.
Fusion receptor
A detailed description of the Tar receptor and LuxQ quorum-sensing receptor can be found in the project description. Since it is believed that the linker region (TM2) is critically for signal transduction two different receptor constructs are designed, referred to as Fusion-1 and Fusion-2. Fusion-1 contains the cytoplasmic domain of Tar and TM2 to the N-terminal of LuxQ (TM2, periplasmic domain, TM1). Fusion-2 contains the cytoplasmic domain and TM2 of Tar and the periplasmic domain to the N-terminal end of LuxQ (periplasmic domain, TM1).
The receptor chimeras will be built in two sequential PCR reactions. In a first PCR the two single fragments containing parts of the coding sequence of LuxQ or Tar will be generated. The reverse primer for LuxQ and the forward primer for Tar have complementary ends, allowing annealing during the second PCR reaction. In addition to the amplified sequences from reaction 1 the forward primer for LuxQ and the reverse primer for Tar will be added to the reaction mix in order to obtain the chimeric receptor sequence.
References
[1] Dong, X.; Stothard, P.; Forsythe, I. J. & Wishart, D. S., PlasMapper: a web server for drawing and auto-annotating plasmid maps, Nucleic Acids Res, Vol. 32, pp. W660-W664, 2004
 "
"