Team:Heidelberg/Notebook/Sensing Group/Notebook/5thweek
From 2008.igem.org
(Difference between revisions)
Chenchenzhu (Talk | contribs) (→Friday, 09/05/2008) |
(→Monday, 09/01/2008) |
||
| Line 125: | Line 125: | ||
== Monday, 09/01/2008 == | == Monday, 09/01/2008 == | ||
* 1.selection for Fusion-1 and Fusion-2 cloning | * 1.selection for Fusion-1 and Fusion-2 cloning | ||
| - | * the DNA of the colonies was extracted with MINIprep, and then screen with test digestion: PstI ( | + | * the DNA of the colonies was extracted with MINIprep, and then screen with test digestion: PstI (NEBuffer 3 + BSA) |
** for Fusion-1: if our insert is there: one band of 6101 bp if no inser: two bands: 4388 + 2213 bp | ** for Fusion-1: if our insert is there: one band of 6101 bp if no inser: two bands: 4388 + 2213 bp | ||
** for Fusion-2: if our insert is there: one band of 6122 bp if no inser: two bands: 4388 + 2213 bp | ** for Fusion-2: if our insert is there: one band of 6122 bp if no inser: two bands: 4388 + 2213 bp | ||
Revision as of 23:39, 26 October 2008


Contents |
Monday, 09/01/2008
- 1.selection for Fusion-1 and Fusion-2 cloning
- the DNA of the colonies was extracted with MINIprep, and then screen with test digestion: PstI (NEBuffer 3 + BSA)
- for Fusion-1: if our insert is there: one band of 6101 bp if no inser: two bands: 4388 + 2213 bp
- for Fusion-2: if our insert is there: one band of 6122 bp if no inser: two bands: 4388 + 2213 bp
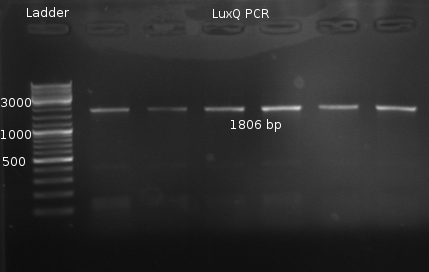
PCR of LuxQ and two fusion protein constructs
- Lyse V. harveyi for 5 min @ 99 °C
- PCR with Phusion (template: supernatend of boiled V. harveyi (a), colony (b))
- 5 min @ 98 °C || 20s @ 98 °C | 40s @ 58 °C | 1min @ 72 °C || 10 min @ 72 °C | 4 °C hold ( 35 cycles )
- 35(34) µl H2O, 10 µl 5x HF, 0.5 µl each primer, 1µl Template, 2µl dNTPs, 1µl Phusion, 0(1) µl DMSO
- PCR with Pfu
- 3 min @ 94 °C || 45s @ 94 °C | 45s @ 58 °C | 2min @ 72 °C || 10 min @ 72 °C | 4 °C hold ( 35 cycles )
- 40(39) µl H2O, 5 µl 10x Buffer, 0.5 µl each primer, 1µl Template, 2µl dNTPs, 1µl Pfu, 0(1) µl DMSO
NO PCR Product
Tuesday, 09/02/2008
- PCR of LuxQ with taq-Polymerase with different DMSO concentrations (0 %, 5 %, 10 %). As template V. harveyi colonies and supernatant of cooked bacteria was used.
No PCR product
Wednesday, 09/03/2008
- PCR of LuxP and LuxQ from Virio harveyi genome with Phusion Mastermix (without any annealing temp). Template: 0.2 µl V. harveyi culture incubated over night in LB(not 20% NaCl LB) at 37 °C.
- result: no PCR product
- PCR of LuxP and LuxQ from Vibrio harveyi genome with Fementas tag Mastermix
- sample with normal Mg concentration (2 µg) and 6 µg Mg concentration
- 5min @94°C ||30 sec @ 94°C | 30 sec @ 58°C | 2 min @ 72°C || 5 min @ 72°C | 4°C hold (30 cycles)
- template is 0.2 ul water washed Vibrio harveyi cells.
- result: see tomorrow (NO PRODUCT)
- incubation of Vibrio harveyi from glycerol stock at 28 over night (medium is 20% NaCl LB)
Thursday, 09/04/2008
- PCR of LuxP and LuxQ from Vibrio hayveyi genome with Tag Mastermix
- template is 0.3 ul water washed Vibrio harveyi culture from 3rd September sample with DMSO(1 ul 100%DMSO) and without DMSO
- Result: only byproduct
Friday, 09/05/2008
- Cloning of LuxQ from day before into DH5a cells with standard protocols
- Plated over weekend @ room temperature
 "
"