Team:KULeuven/Modeling
From 2008.igem.org
(Difference between revisions)
m (→Cell Death) |
(→Inverter) |
||
| (47 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
{{:Team:KULeuven/Tools/Navigation_Bar}} | {{:Team:KULeuven/Tools/Navigation_Bar}} | ||
| - | + | = Modeling = | |
| - | + | * [https://static.igem.org/mediawiki/2008/1/19/Numbers.xls Numbers.xls] | |
| - | + | ||
| - | * [ | + | |
* [http://www.nature.com/nature/journal/v438/n7067/full/nature04405.html Synthetic biology: Engineering Escherichia coli to see light] | * [http://www.nature.com/nature/journal/v438/n7067/full/nature04405.html Synthetic biology: Engineering Escherichia coli to see light] | ||
| - | ==CellDesigner== | + | == CellDesigner == |
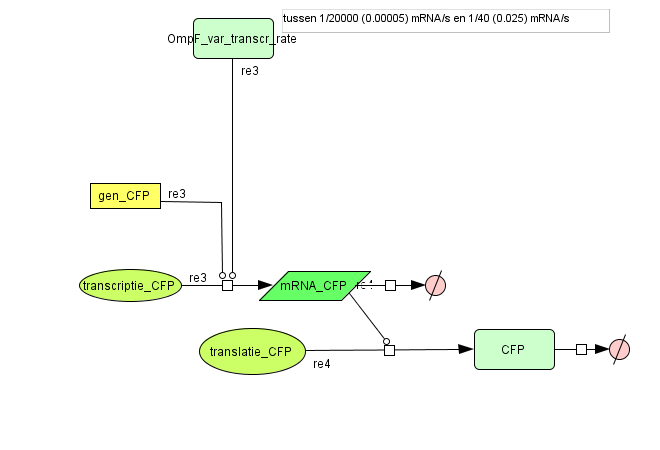
| - | ===[https://static.igem.org/mediawiki/2008/d/d9/Output_v2.zip Output] === | + | === [https://static.igem.org/mediawiki/2008/d/d9/Output_v2.zip Output] === |
| + | |||
| + | [[Image:output_v2.png|500px|center|Output]] | ||
| + | |||
| + | {| border="1" | ||
| + | |+ Parameter values Output | ||
| + | ! Name !! Value !! Reference | ||
| + | |- | ||
| + | ! Degradation | ||
| + | | || | ||
| + | |- | ||
| + | | CFP (protein) || 1.05E-4 /s || [https://2007.igem.org/ETHZ/Parameters link] | ||
| + | |- | ||
| + | | CFP (mRNA) || 0.0023 /s || [http://www.pubmedcentral.nih.gov/picrender.fcgi?artid=124983&blobtype=pdf link] | ||
| + | |- | ||
| + | ! Transcription Rate | ||
| + | |- | ||
| + | | CFP || OmpF dependent || | ||
| + | |- | ||
| + | ! Translation Rate | ||
| + | |- | ||
| + | | CFP || 0.167 /s || Based on effenciency RBS | ||
| + | |} | ||
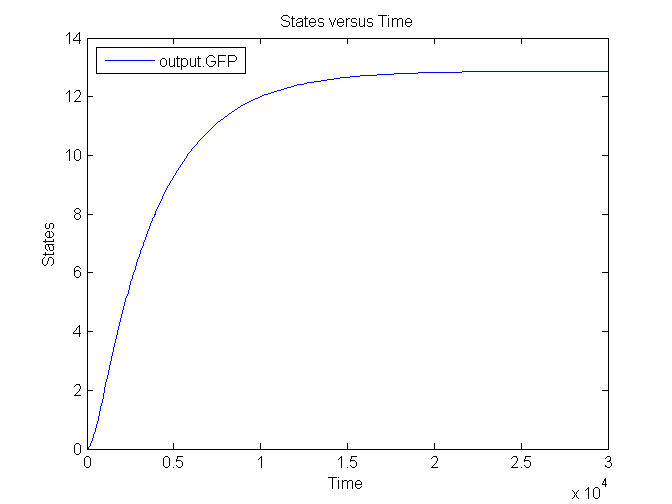
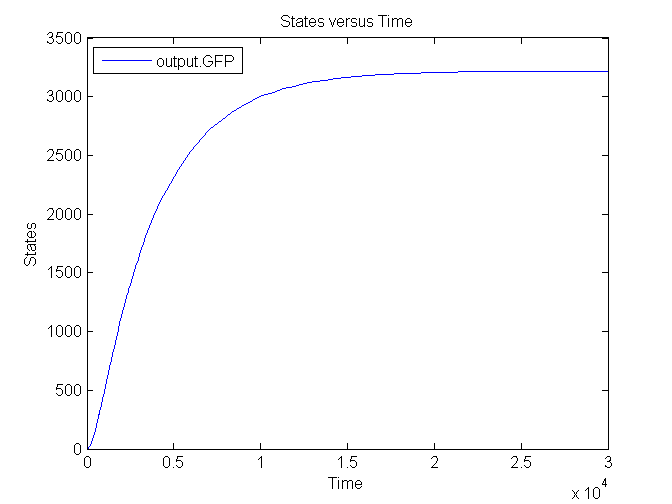
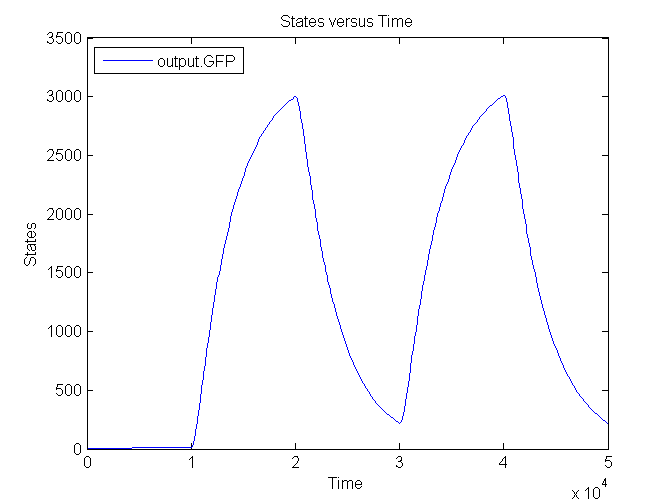
| - | [[Image: | + | [[Image:Sim_output_1.png|500px|center|Output]] |
| + | [[Image:Sim_output_2.png|500px|center|Output]] | ||
| + | [[Image:Sim_output_3.png|500px|center|Output]] | ||
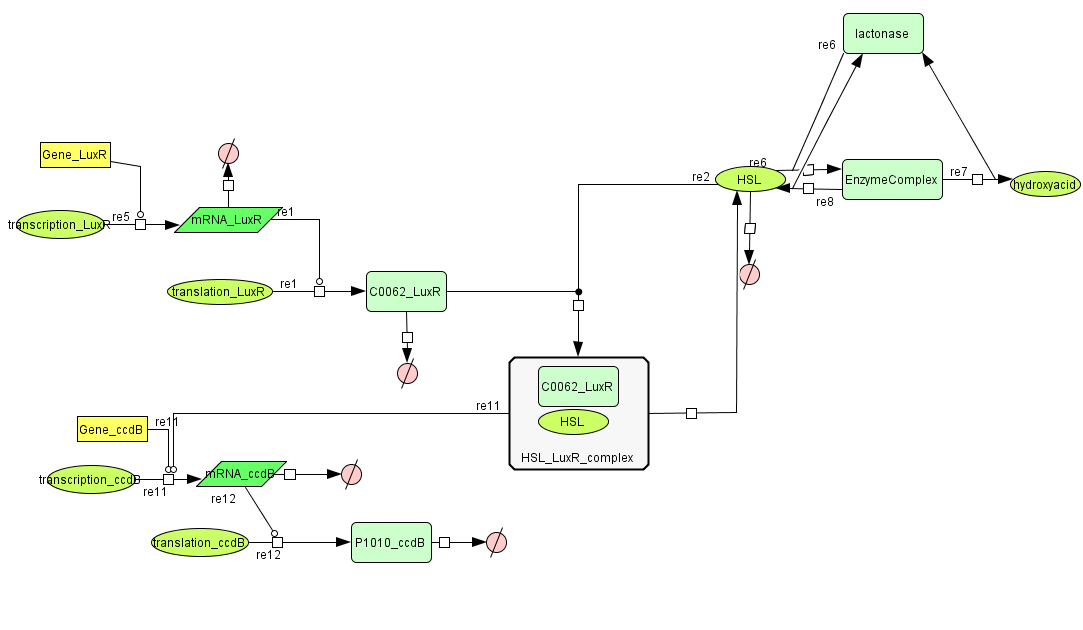
===[https://static.igem.org/mediawiki/2008/4/47/Celldeath.zip Cell Death]=== | ===[https://static.igem.org/mediawiki/2008/4/47/Celldeath.zip Cell Death]=== | ||
| - | [[Image:celldeath.png| | + | [[Image:celldeath.png|800px|center|Cell Death]] |
{| border="1" | {| border="1" | ||
| Line 28: | Line 50: | ||
| LuxR (protein) || 0.0010 /s || | | LuxR (protein) || 0.0010 /s || | ||
|- | |- | ||
| - | | LuxR (mRNA) || 0.00227 /s || | + | | LuxR (mRNA) || 0.00227 /s || [http://www.pubmedcentral.nih.gov/picrender.fcgi?artid=124983&blobtype=pdf link] |
|- | |- | ||
| CcdB (protein) || 7.7E-5 /s || [http://www.ncbi.nlm.nih.gov/pubmed/8022284?dopt=abstract link] | | CcdB (protein) || 7.7E-5 /s || [http://www.ncbi.nlm.nih.gov/pubmed/8022284?dopt=abstract link] | ||
|- | |- | ||
| - | | CcdB (mRNA)|| 0.00231 /s || | + | | CcdB (mRNA)|| 0.00231 /s || [http://www.pubmedcentral.nih.gov/picrender.fcgi?artid=124983&blobtype=pdf link] |
|- | |- | ||
| - | | HSL || 1.02E-6/s || | + | | HSL || 1.02E-6/s || [http://aem.asm.org/cgi/content/abstract/71/3/1291 link] |
|- | |- | ||
! Dissociation Rate | ! Dissociation Rate | ||
|- | |- | ||
| - | | HSL | + | | k<sub>a</sub> (HSL+LuxR) || 1E6 /s || chosen to be relatively (to the other rate constants) high and such that K<sub>d</sub> equals 1E-6 |
| + | |- | ||
| + | | k<sub>d</sub> (HSL+LuxR) || 1 /s || | ||
|- | |- | ||
! Dissociation Constant | ! Dissociation Constant | ||
|- | |- | ||
| - | | Binding LuxR on LuxPromotor || 1E-9 M/L || [http:// | + | | HSL-LuxR || 1E-6 M/L || [http://jb.asm.org/cgi/content/full/189/11/4127?view=long&pmid=17400743 link] |
| + | |- | ||
| + | | Binding LuxR on LuxPromotor || 1E-9 M/L || [http://jb.asm.org/cgi/content/full/189/11/4127?view=long&pmid=17400743 link] | ||
| + | |- | ||
| + | ! Transcription Rate | ||
| + | |- | ||
| + | | LuxR (constitutive promotor) || 0.025 mRNA/s || see sections on Constitutive Promotors & E. coli transcription Rates | ||
| + | |- | ||
| + | | CcdB (LuxR repressor) || k_1*K_m^n/(K_m^n + [LuxR]^n) = 0.025*1E-9/(1E-9^1+[LuxR]) || | ||
| + | |- | ||
|} | |} | ||
| Line 49: | Line 82: | ||
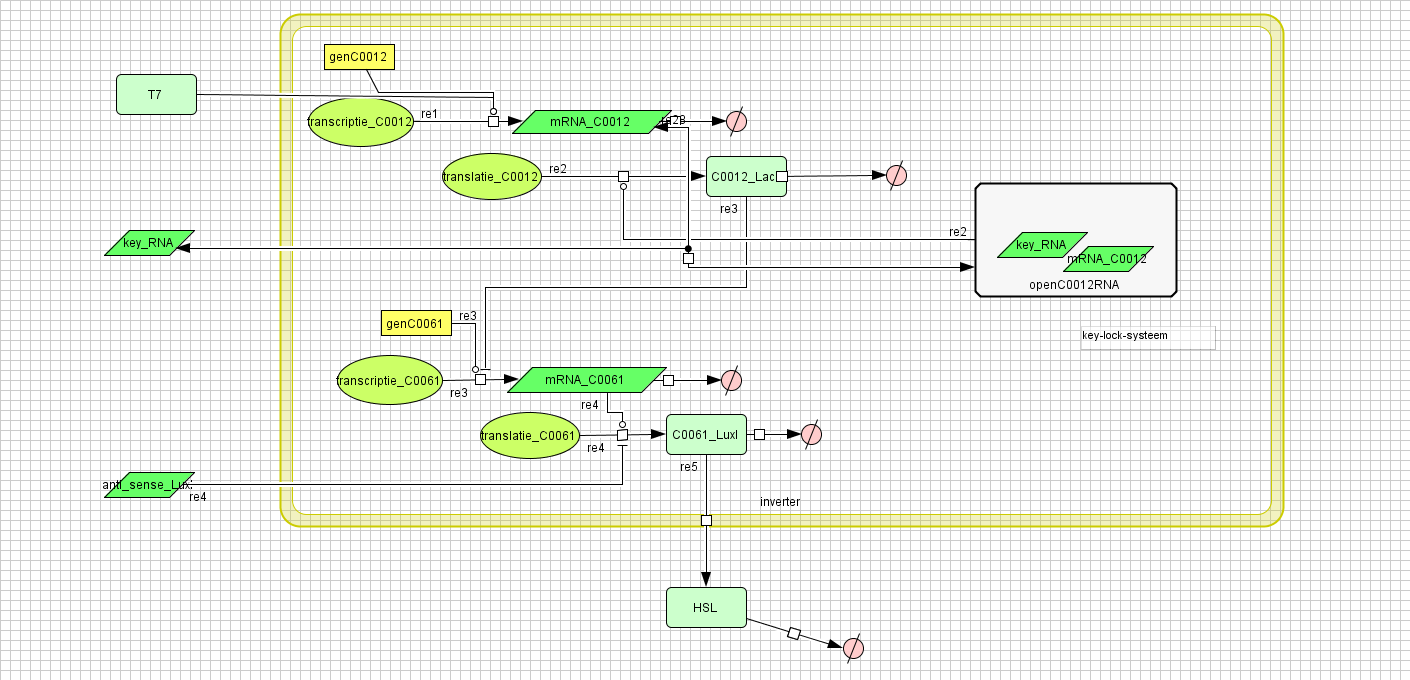
[[Image:inverter_v4.png|400px|center|Inverter]] | [[Image:inverter_v4.png|400px|center|Inverter]] | ||
| - | + | {| border="1" | |
| - | + | |+ Parameter values Inverter | |
| - | + | ! Name !! Value !! Reference | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | {| | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
|- | |- | ||
| - | + | ! Degradation | |
| - | | | + | | || |
|- | |- | ||
| - | | | + | | LuxI (protein) || 0.00231 /s || |
| - | | | + | |
|- | |- | ||
| - | | | + | | LuxI (mRNA) || 0.0025 /s || [http://www.pubmedcentral.nih.gov/picrender.fcgi?artid=124983&blobtype=pdf link] |
| - | | | + | |
| - | | | + | |
| - | | | + | |
| - | + | ||
| - | + | ||
|- | |- | ||
| - | | | + | | LuxI (complex) || 0.00125 /s || |
| - | | | + | |
|- | |- | ||
| - | | | + | | LacI (protein) || 0.00231 /s || |
| - | | | + | |
|- | |- | ||
| - | | | + | | LacI (gesloten mRNA) || 0.0046209812 /s || |
| - | | | + | |
|- | |- | ||
| - | | | + | | LacI (open mRNA) || 0.0023104906 /s || |
| - | | | + | |
|- | |- | ||
| - | | | + | | open mRNA LacI complex || 0.0023104906 /s || |
| - | | | + | |
|- | |- | ||
| - | | | + | | HSL || 1.02E-6 /s || [http://aem.asm.org/cgi/content/abstract/71/3/1291 link] |
| - | | | + | |
|- | |- | ||
| - | + | ! Dissociation Rate | |
| - | + | ||
|- | |- | ||
| - | + | ! Transcription Rate | |
| - | + | ||
|- | |- | ||
| - | | | + | | LacI || 0.0011 /s || |
| - | | | + | |
|- | |- | ||
| - | | | + | | LuxI || 0.0025 /s || |
| - | | | + | |
|- | |- | ||
| - | | | + | | K_m || 1.0E-10 /s || |
| - | | | + | |
| - | |- | + | |
| - | | | + | |
| - | + | ||
| - | | | + | |
|- | |- | ||
| - | + | ! Translation Rate | |
| - | + | ||
|- | |- | ||
| - | | | + | | LuxI (RBS B0034) || 0.5 /s || |
| - | | | + | |
|- | |- | ||
| - | | | + | | LacI (RBS ?) || 0.16667 /s || |
| - | | | + | |
|- | |- | ||
| - | + | ! Equilibrium Constant | |
| - | + | ||
|- | |- | ||
| - | | | + | | closed and open mRNA LacI || 0.015 || |
| - | | | + | |
|- | |- | ||
| - | | | + | | closed and complex mRNA LacI || 0.0212 /M || |
| - | | | + | |
|- | |- | ||
| - | + | ! Hill coëfficiënt | |
| - | + | ||
|- | |- | ||
| - | | | + | | n || 2 || |
| - | | | + | |
| - | | | + | |
| - | | | + | |
| - | + | ||
| - | | | + | |
|- | |- | ||
| - | + | ! Others | |
| - | + | ||
|- | |- | ||
| - | | | + | | LuxI --> HSL || 0.16 || |
| - | | | + | |
|- | |- | ||
| - | | | + | | LuxI (mRNA) + anti sense <-> complex LuxI || 4.22E14 || |
| - | | | + | |
|- | |- | ||
| - | | | + | | |
| - | | | + | |} |
| + | |||
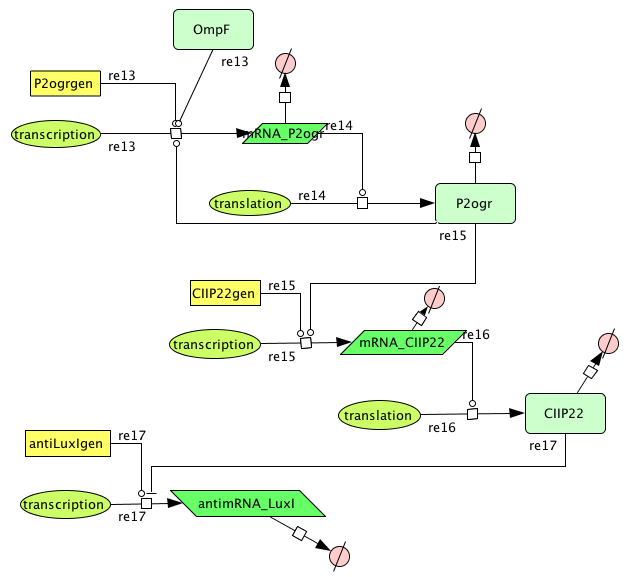
| + | ===[https://static.igem.org/mediawiki/2008/0/05/Memory.zip Memory]=== | ||
| + | |||
| + | [[Image:Memory.png|400px|center|Memory]] | ||
| + | |||
| + | {| border="1" | ||
| + | |+ Parameter values Memory | ||
| + | ! Name !! Value !! Reference | ||
|- | |- | ||
| - | + | ! Degradation | |
| - | + | ||
|- | |- | ||
| - | | | + | | P2ogr (protein) || 0.002265 /s || |
| - | | | + | |
|- | |- | ||
| - | | | + | | P2ogr (mRNA) || 0.002265 /s || [http://www.pubmedcentral.nih.gov/picrender.fcgi?artid=124983&blobtype=pdf link] |
| - | | | + | |
|- | |- | ||
| - | | | + | | P22CII (protein) || 0.002311 /s || |
| - | | | + | |
|- | |- | ||
| - | | | + | | P22CII (mRNA)|| 0,0022651 /s || [http://www.pubmedcentral.nih.gov/picrender.fcgi?artid=124983&blobtype=pdf link] |
| - | | | + | |
|- | |- | ||
| - | | | + | | antimRNA_luxI || 0.0045303/s || |
| - | | | + | |
|- | |- | ||
| - | + | ! Transcription rate | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
|- | |- | ||
| - | | | + | | P2ogr || 0.0125 || |
| - | | | + | |
|- | |- | ||
| - | | | + | | P22CII || 0.0125 || |
| - | | | + | |
|- | |- | ||
| - | | | + | | AntimRNA || 0.0094 || |
| - | | | + | |
|- | |- | ||
| - | | | + | ! Dissociation Constant |
| - | | | + | |- |
| + | | P2-ogr || 4.2156 M || [http://www.jbc.org/cgi/content/abstract/258/17/10536?maxtoshow=&HITS=10&hits=10&RESULTFORMAT=&fulltext=P22+c2+repressor&andorexactfulltext=and&searchid=1&FIRSTINDEX=0&sortspec=relevance&volume=258&resourcetype=HWCIT link] | ||
| + | |- | ||
| + | | R0053-P22CII || 0.1099 M || | ||
| + | |- | ||
| + | ! Hill coëfficiënt | ||
| + | |- | ||
| + | | n || 2 || | ||
| + | |- | ||
| + | |} | ||
| + | |||
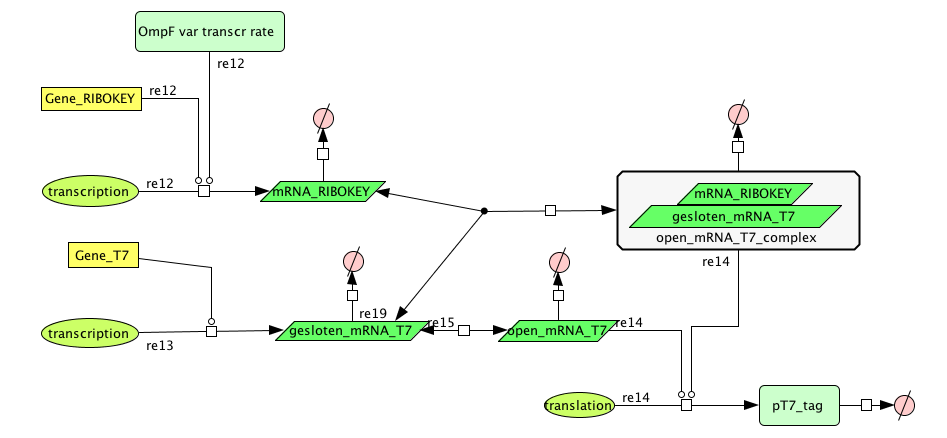
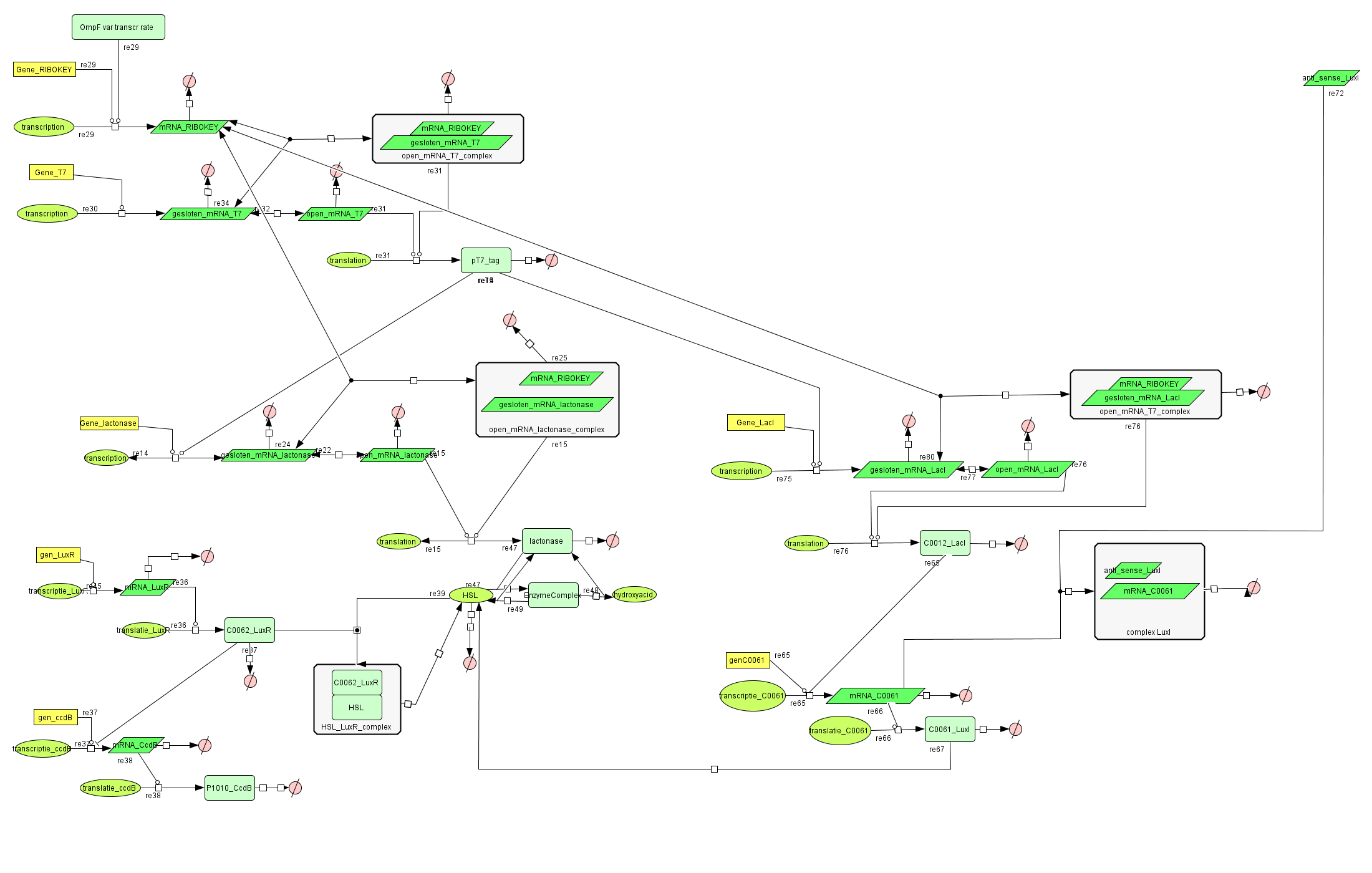
| + | ===[https://static.igem.org/mediawiki/2008/8/8b/Filter_Final.zip Filter]=== | ||
| + | |||
| + | [[Image:FilterCellD.png|800px|center|filter]] | ||
| + | |||
| + | {| border="1" | ||
| + | |+ Parameter values Filter | ||
| + | ! Name !! Value !! Reference | ||
| + | |- | ||
| + | ! Degradation | ||
| + | | || | ||
| + | |- | ||
| + | | pT7_tag(protein) || 0.00155/s || | ||
| + | |- | ||
| + | | mRNA_RIBOKEY (RNA) || 0.00462 /s ||[http://www.pubmedcentral.nih.gov/picrender.fcgi?artid=124983&blobtype=pdf link] | ||
|- | |- | ||
| - | | | + | |gesloten_mRNA_T7 (RNA) || 0.00462 /s ||[http://www.pubmedcentral.nih.gov/picrender.fcgi?artid=124983&blobtype=pdf link] |
| - | | | + | |
|- | |- | ||
| - | | | + | |open_mRNA_T7 (RNA) ||0.00231 / s||[http://www.pubmedcentral.nih.gov/picrender.fcgi?artid=124983&blobtype=pdf link] |
| - | | | + | |
|- | |- | ||
| - | | | + | |open_mRNA_T7_complex (RNA) || 0.00231 /s||[http://www.pubmedcentral.nih.gov/picrender.fcgi?artid=124983&blobtype=pdf link] |
| - | | | + | |
|- | |- | ||
| - | + | ! Equilibrium Constant | |
| - | + | ||
|- | |- | ||
| - | | | + | | closed and open mRNA || 0,015|| |
| - | | | + | |
|- | |- | ||
| - | | | + | | closed and key complex mRNA || 0.0212 /M || |
| - | | | + | |
|- | |- | ||
| - | + | ! Transcription Rate | |
| - | + | ||
|- | |- | ||
| - | | | + | | mRNA_RIBOKEY || Input dependent || |
| - | | | + | |
| - | | | + | |
| - | | | + | |
|- | |- | ||
| - | | | + | |mRNA_T7 || 0,0011 mRNA/s || |
| - | | | + | |
|} | |} | ||
| + | |||
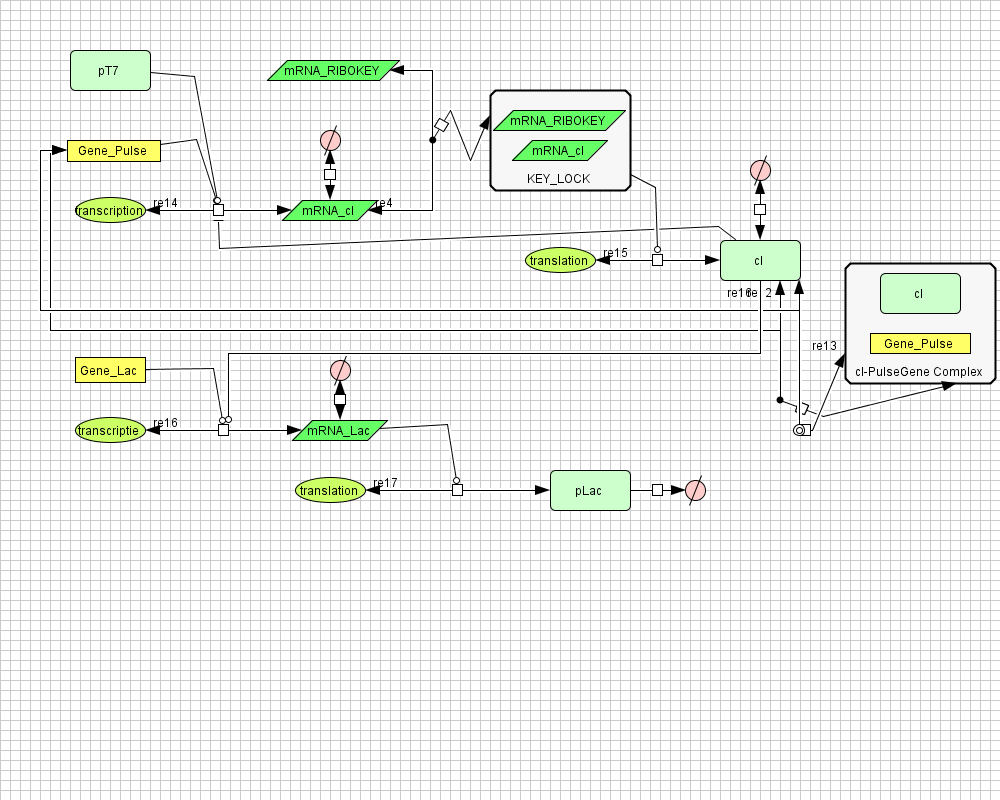
| + | ===[https://static.igem.org/mediawiki/2008/8/8c/PulseGenerator.zip Pulse Generator]=== | ||
| + | |||
| + | [[Image:PulseGenerator.png|400px|center|Pulse Generator]] | ||
| + | |||
| + | ===Constant Lactonase production=== | ||
| + | |||
| + | ===[https://static.igem.org/mediawiki/2008/c/c0/FilterLactonaseCelldeathInverter.zip Partial Model]=== | ||
| + | |||
| + | [[Image:Model.png|950px|center|Model]] | ||
| + | |||
| + | ==Kinetic Constants== | ||
===ETHZ list of parameters=== | ===ETHZ list of parameters=== | ||
| - | * [ | + | * [https://2007.igem.org/ETHZ/Parameters ETHZ list of parameters] |
===mRNA decay=== | ===mRNA decay=== | ||
*[http://www.pubmedcentral.nih.gov/picrender.fcgi?artid=124983&blobtype=pdf Global analysis of mRNA decay and abundance in Escherichia coli at single-gene resolution using two-color fluorescent DNA microarrays] | *[http://www.pubmedcentral.nih.gov/picrender.fcgi?artid=124983&blobtype=pdf Global analysis of mRNA decay and abundance in Escherichia coli at single-gene resolution using two-color fluorescent DNA microarrays] | ||
| Line 236: | Line 235: | ||
*[http://pubs.acs.org/cgi-bin/article.cgi/bichaw/2002/41/i11/pdf/bi0158472.pdf Kinetic and Thermodynamic Basis of Promoter Strength: Multiple Steps of Transcription Initiation by T7 RNA Polymerase Are Modulated by the Promoter Sequence] | *[http://pubs.acs.org/cgi-bin/article.cgi/bichaw/2002/41/i11/pdf/bi0158472.pdf Kinetic and Thermodynamic Basis of Promoter Strength: Multiple Steps of Transcription Initiation by T7 RNA Polymerase Are Modulated by the Promoter Sequence] | ||
*[http://www.openwetware.org/wiki/IGEM:Tsinghua/2007/Projects/RAP data T7 team Tsinghua 2007] | *[http://www.openwetware.org/wiki/IGEM:Tsinghua/2007/Projects/RAP data T7 team Tsinghua 2007] | ||
| + | *[http://www.jbc.org/cgi/reprint/281/47/35677 Transient State Kinetics of Transcription Elongation by T7 RNA Polymerase] | ||
===LacI - LuxI=== | ===LacI - LuxI=== | ||
| Line 246: | Line 246: | ||
*[http://bioinformatics.oxfordjournals.org/cgi/content/full/21/11/2722#E11 Noise-induced cooperative behavior in a multicell system] | *[http://bioinformatics.oxfordjournals.org/cgi/content/full/21/11/2722#E11 Noise-induced cooperative behavior in a multicell system] | ||
| - | ===HSL | + | *[http://www.pnas.org/content/93/18/9505 LuxI kinetics] |
| + | |||
| + | ===HSL stuff=== | ||
*[http://aem.asm.org/cgi/content/abstract/71/3/1291 Rapid Acyl-Homoserine Lactone Quorum Signal Biodegradation in Diverse Soils] Fig 6: half-life 185 h ==> decay rate 1.02 * 10^-6 s^-1 ==> 0.00889 nM/h | *[http://aem.asm.org/cgi/content/abstract/71/3/1291 Rapid Acyl-Homoserine Lactone Quorum Signal Biodegradation in Diverse Soils] Fig 6: half-life 185 h ==> decay rate 1.02 * 10^-6 s^-1 ==> 0.00889 nM/h | ||
| + | |||
| + | *[http://www.sciencedirect.com/science?_ob=ArticleURL&_udi=B6WK7-457D0X9-9&_user=877992&_rdoc=1&_fmt=&_orig=search&_sort=d&view=c&_acct=C000047079&_version=1&_urlVersion=0&_userid=877992&md5=36d07f326b2c2e55c4de05a1ab426e09 Kinetics of the AHL Regulatory System in a Model Biofilm System] | ||
| + | |||
| + | *[http://www.pnas.org/content/96/8/4360.full HSL lactone synthesis kinetics by a LuxI-related enzyme] | ||
| + | |||
| + | *[http://www.jbc.org/cgi/content/full/279/14/13645#TBL2 Lactonase mediated HSL degradation (hydrolysis)] | ||
===OmpR, OmpF=== | ===OmpR, OmpF=== | ||
| Line 259: | Line 267: | ||
===Psid met P2ogr promotor=== | ===Psid met P2ogr promotor=== | ||
| - | *[ | + | *[https://2007.igem.org/Cambridge/Amplifier_project Cambridge amplifier project, combinatie van verschillende promotoren met verschillende activatoren, experimenteel amplification factor bepaald met GFP reporter] |
| + | |||
| + | *[http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=178405 P<sub>sid</sub> promotor with different activators] | ||
===Constitutive promoters=== | ===Constitutive promoters=== | ||
| - | *Estimated transcription rate for J23105:[ | + | *Estimated transcription rate for J23105:[https://2007.igem.org/title=ETHZ/Parameters] |
*Scale other transcription rate with table in parts registry. | *Scale other transcription rate with table in parts registry. | ||
* Estimate the rate of transcription from a constitutive promotor family member. | * Estimate the rate of transcription from a constitutive promotor family member. | ||
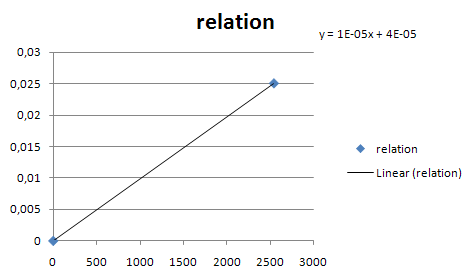
| - | [[Image:Const_pro_strength.png|center|400px]] | + | [[Image:Const_pro_strength.png|center|400px|thumb|Estimates for the rate of transcription from the constitutive promotor family members. X is the GFP fluorescence in arbitrary units according to the Registry. Y is the number of mRNA's produced per second from that promotor]] |
===E. coli transcription rates=== | ===E. coli transcription rates=== | ||
[http://www.fasebj.org/cgi/content/summary/20/10/1721 Paper about the calculated transcription rates for every E.coli ORF] <br> | [http://www.fasebj.org/cgi/content/summary/20/10/1721 Paper about the calculated transcription rates for every E.coli ORF] <br> | ||
[http://users.path.ox.ac.uk/~pcook/data/catalogs.html Tables with the calculated transcription rates for every E.coli ORF] | [http://users.path.ox.ac.uk/~pcook/data/catalogs.html Tables with the calculated transcription rates for every E.coli ORF] | ||
Latest revision as of 15:17, 22 July 2008
Result:
To have a dropdown for your own team copy over all of the content below. It might be needed to adjust a thing or two. Comments have been added throughout the code what must be changed.
Inspirational websites:
- [http://javascript-array.com/scripts/simple_drop_down_menu/ Most basic dropdown menu, with no submenu's]
- [http://jquery.com/ The javascript library used to add effects and optimise the dropdown]
- [http://www.tyssendesign.com.au/articles/css/centering-a-dropdown-menu/ How to center a ul list effectively]
The dropdown has been created and developed by the KULeuven team.
<html>
<style type="text/css">
#content {z-index:4;}
#ddwrapper * {z-index:8 !important;}
div#ddwrapper {
margin:0;
padding:0;
height:28px;
width:945px; /* change to adjust imperfections in width */
}
div#ddnav {
margin:0 auto; /* needed to center the dropdown */
padding:0;
top:5px;
/* width: 965px */
visibility:hidden; /* dropdown is hidden until properly initalised by javascript */
}
div#ddtoggle {
margin:0;
position:fixed;
right:2px;
top:15px;
height:10px;
width:10px;
z-index:100;
}
#ddnav ul {
display:table-row; /* works only for firefox, later adjusted by javascript for IE */
margin:0 auto;
padding:0;
}
#ddnav ul li {
display:table-cell; /* works only for firefox, later adjusted by javascript for IE */
list-style:none;
margin:0;
padding:0 !important;
border-right:1px solid #FFF; /* creates illusion of spacing between tabs */
}
#ddnav ul li:last-child{border-right:none;}
#ddnav a{
display:block;
margin:0;
padding:4px 14px; /* play with dimensions for size of tabs */
background-color:#075A90; /* background color for tabs */
color:#FFF !important; /* font color for text in tabs */
text-align:center; /* aligning for text in tabs */
text-decoration:none !important;
font:bold 10pt Trebuchet MS; /* font properties for text in tabs */
outline:0;
}
#ddnav ul li a:hover {background-color:#99CCFF;}/* background color for tabs on mouseover */
#ddnav li a:active {outline:none;} /* remove standard dotted border for links when clicked (IE) */
#ddnav li a:focus {-moz-outline-style:none;} /* remove standard dotted border for links when clicked (FF) */
#ddnav div {
display:none;
position:absolute;
width:9em;
background-color:#000; /* bug solution, do not change ! */
border:1px solid #5970B2; /* border color for dropdown menus */
opacity:0.9; /* transparancy of the dropdown menus (FF) */
filter:alpha(opacity=90); /* transparancy of the dropdown menus (IE) */
}
#ddnav div a {
display:block;
padding:5px 10px; /* play with dimensions of block element in dropdown menus */
position:relative;
font:normal 8pt arial; /* font properties for text in dropdown menus */
text-align:left; /* aligning of text in dropdown menus */
cursor:pointer;
}
#ddnav div a:hover, #ddnav span a:hover {color:#000 !important;} /* text color on mouseover */
#ddnav span div {
position:relative;
border:none;
border-bottom:2px groove #5970B2; /* separator for submenus, groove does not work in FF */
opacity:1.0; /* avoid stacking transparancy for submenus (FF) */
filter:alpha(opacity=100); /* avoid stacking transparancy for submenus (IE) */
}
/* may want to upload the following pictures to a new location */
.expand {background: url('https://static.igem.org/mediawiki/2008/e/ef/Icon-expand.png') no-repeat 95% 50%;}
.collapse {background: url('https://static.igem.org/mediawiki/2008/c/cd/Icon-collapse.png') no-repeat 95% 50%;}
.docked {background: #99ccff url("https://static.igem.org/mediawiki/2008/6/62/Ddnavundock.png") no-repeat 50% 50%;}
.undocked {background: #99ccff url("https://static.igem.org/mediawiki/2008/e/e4/Ddnavdock.png") no-repeat 50% 50%;}
</style>
<!-- IMPORTANT: save following script under a personalized webspace or download the library at www.jquery.com -->
<script type="text/javascript" src="http://student.kuleuven.be/~s0173901/wiki/js/jquery.js"></script>
<script type="text/javascript">
function ddnav() {
$('#ddnav ul li').hover(
function () {
$(this).find('div:first').css('display','block');},
function () {
$(this).find('div:first').css('display','none');}
);
}
function ddnavsub() {
$('#ddnav span > a').toggle(
function () {
$(this).removeClass("#ddnav expand").addClass("#ddnav collapse");
$(this).parent().find('div:first').slideDown('fast');
$(this).hover(
function (){$(this).css('background-color','#99AAFF');},
function (){$(this).css('background-color','#99AAFF');});},
function () {
$(this).removeClass("#ddnav collapse").addClass("#ddnav expand");
$(this).parent().find('div:first').css('display','none');
$(this).hover(
function (){$(this).css('background-color','#99CCFF');},
function (){$(this).css('background-color','#075A90');});}
).addClass("#ddnav expand");
}
/* If you wish to omit the docking feature, remove following function ddtoggle() */
function ddtoggle() {
$('#ddtoggle').toggle(
function () {
$(this).removeClass('undocked').addClass('docked');
$('#ddnav').css('position','fixed');},
function () {
$(this).removeClass('docked').addClass('undocked');
$('#ddnav').css('position','static');}
);
}
function ddalign() {
var _windowWidth = $(window).width();
var _leftOffset = (_windowWidth - 965)/2;
$('div#ddnav').css('left',_leftOffset);
}
function ddmsie() {
$('#ddnav a').hover(
function () {$(this).css('background-color','#99ccff');},
function () {$(this).css('background-color','#075a90');}
);
/* toggle doesn't work yet */
$('#ddtoggle').css('display','none');
$('#ddnav ul').css('display','inline-block');
$('#ddnav ul li').css('display','inline');
$('#ddnav ul li').css('position','relative');
$('#ddnav ul li>a').css('display','inline-block');
$('#ddnav ul li>a').css('margin-right','-4px');
$('#ddnav div').css('left','0');
$('#ddnav div').css('top','100%');
$('#ddnav span div').css('top','0');
}
function ddmozilla() {
ddtoggle();
$(window).bind('resize', function() {ddalign();});
}
$(function () {
ddnav();
ddnavsub();
if(jQuery.browser.msie) ddmsie();
if(jQuery.browser.mozilla) ddmozilla();
$('#ddnav').css('visibility','visible');
});
</script>
<!-- If you wish to omit the docking feature omit following line (div with id ddtoggle) -->
<div id="ddtoggle" class="undocked"></div>
<div id="ddwrapper">
<!-- Here the actual links are defined, simply replace with your own links in the appropriate sections -->
<div id="ddnav" align="center">
<ul>
<li>
<a href="https://2008.igem.org/Team:KULeuven">Home</a>
</li>
<li>
<a>The Team</a>
<div>
<a href="https://2008.igem.org/Team:KULeuven/Team/LabsandGroups">Research Labs and Groups</a>
<a href="https://2008.igem.org/Team:KULeuven/Team/Students">Students</a>
<a href="https://2008.igem.org/Team:KULeuven/Team/Instructors">Instructors</a>
<a href="https://2008.igem.org/Team:KULeuven/Team/Advisors">Advisors</a>
<a href="https://2008.igem.org/Team:KULeuven/Team/Pictures">Pictures</a>
</div>
</li>
<li>
<a>The Project</a>
<div>
<a href="https://2008.igem.org/Team:KULeuven/Project">Summary</a>
<span>
<a>Components</a>
<div>
<a href="https://2008.igem.org/Team:KULeuven/Project/Input">Input</a>
<a href="https://2008.igem.org/Team:KULeuven/Project/Output">Output</a>
<a href="https://2008.igem.org/Team:KULeuven/Project/Filter">Filter</a>
<a href="https://2008.igem.org/Team:KULeuven/Project/Inverter">Invertimer</a>
<a href="https://2008.igem.org/Team:KULeuven/Project/Reset">Reset</a>
<a href="https://2008.igem.org/Team:KULeuven/Project/CellDeath">Cell Death</a>
<a href="https://2008.igem.org/Team:KULeuven/Project/Memory">Memory</a>
</div>
</span>
<a href="https://2008.igem.org/Team:KULeuven/Evaluation">End Evaluation</a>
<a href="https://2008.igem.org/Team:KULeuven/Literature">Literature</a>
<a href="https://2008.igem.org/Team:KULeuven/Brainstorm">Brainstorm</a>
</div>
</li>
<li>
<a>Ethics</a>
<div>
</div>
</li>
<li>
<a>Submitted Parts</a>
<div>
<a href="https://2008.igem.org/Team:KULeuven/Parts">Listing</a>
<a href="http://partsregistry.org/cgi/partsdb/pgroup.cgi?pgroup=iGEM2008&group=KULeuven">Sandbox</a>
</div>
</li>
<li>
<a>Modeling</a>
<div>
<a href="https://2008.igem.org/Team:KULeuven/Model/Overview">Overview</a>
<a href="https://2008.igem.org/Team:KULeuven/Model/KineticConstants">Kinetic Constants</a>
<span>
<a>Components</a>
<div>
<a href="https://2008.igem.org/Team:KULeuven/Model/Output">Output</a>
<a href="https://2008.igem.org/Team:KULeuven/Model/Filter">Filter</a>
<a href="https://2008.igem.org/Team:KULeuven/Model/Inverter">Invertimer</a>
<a href="https://2008.igem.org/Team:KULeuven/Model/Reset">Reset</a>
<a href="https://2008.igem.org/Team:KULeuven/Model/CellDeath">Cell Death</a>
<a href="https://2008.igem.org/Team:KULeuven/Model/Memory">Memory</a>
</div>
</span>
<a href="https://2008.igem.org/Team:KULeuven/Model/FullModel">Full Model</a>
<a href="https://2008.igem.org/Team:KULeuven/Model/Sensitivity">Sensitivity Analysis</a>
<a href="https://2008.igem.org/Team:KULeuven/Model/MultiCell">Multi-cell Model</a>
<a href="https://2008.igem.org/Team:KULeuven/Model/Diffusion">Diffusion</a>
</div>
</li>
<li>
<a>Data Analysis</a>
<div>
<a href="https://2008.igem.org/Team:KULeuven/Data/Overview">Overview</a>
<span>
<a>New Parts</a>
<div>
<a href="https://2008.igem.org/Team:KULeuven/Data/GFP">GFP (LVA-tag)</a>
<a href="https://2008.igem.org/Team:KULeuven/Data/T7">T7 (UmuD-tag)</a>
<a href="https://2008.igem.org/Team:KULeuven/Data/Antisense">Antisense LuxI</a>
<a href="https://2008.igem.org/Team:KULeuven/Data/ccdB">Celldeath (ccdB)</a>
<a href="https://2008.igem.org/Team:KULeuven/Data/HybridProm">Hybrid Promotor</a>
</div>
</span>
<span>
<a>Components</a>
<div>
<a href="https://2008.igem.org/Team:KULeuven/Data/Input">Input</a>
<a href="https://2008.igem.org/Team:KULeuven/Data/Output">Output</a>
<a href="https://2008.igem.org/Team:KULeuven/Data/Filter">Filter</a>
<a href="https://2008.igem.org/Team:KULeuven/Data/Inverter">Invertimer</a>
<a href="https://2008.igem.org/Team:KULeuven/Data/Reset">Reset</a>
<a href="https://2008.igem.org/Team:KULeuven/Data/CellDeath">Cell Death</a>
<a href="https://2008.igem.org/Team:KULeuven/Data/Memory">Memory</a>
</div>
</span>
<a href="https://2008.igem.org/Team:KULeuven/Data/FullModel">Full Model</a>
</div>
</li>
<li>
<a>Software</a>
<div>
<a href="https://2008.igem.org/Team:KULeuven/Software/MultiCell">Multi-cell Toolbox</a>
<a href="https://2008.igem.org/Team:KULeuven/Software/Simbiology2LaTeX">Simbiology2LaTeX Toolbox</a>
</div>
</li>
<li>
<a>Notebook</a>
<div>
<a href="https://2008.igem.org/Team:KULeuven/Meeting_Calendar">Calendar</a>
<a href="https://2008.igem.org/Team:KULeuven/SummerHolidays">Summer Holidays</a>
<span>
<a>Reports</a>
<div>
<a href="https://2008.igem.org/Team:KULeuven/Meeting Reports">Daily</a>
<a href="https://2008.igem.org/Team:KULeuven/Weekly Meetings">Weekly</a>
</div>
</span>
<span>
<a>Lab Data</a>
<div>
<a href="https://2008.igem.org/Team:KULeuven/Freezer">Freezer</a>
<a href="https://2008.igem.org/Team:KULeuven/Primers">Primers</a>
<a href="https://2008.igem.org/Team:KULeuven/Ligation">Ligation</a>
</div>
</span>
<a href="https://2008.igem.org/Team:KULeuven/Tools">Tools</a>
<a href="https://2008.igem.org/Team:KULeuven/Press">Press</a>
<a href="https://2008.igem.org/Team:KULeuven/Guestbook">Guestbook</a>
</div>
</li>
</ul>
</div>
</div>
</html>
Contents |
Modeling
- [http://www.nature.com/nature/journal/v438/n7067/full/nature04405.html Synthetic biology: Engineering Escherichia coli to see light]
CellDesigner
Output
| Name | Value | Reference |
|---|---|---|
| Degradation | ||
| CFP (protein) | 1.05E-4 /s | link |
| CFP (mRNA) | 0.0023 /s | [http://www.pubmedcentral.nih.gov/picrender.fcgi?artid=124983&blobtype=pdf link] |
| Transcription Rate | ||
| CFP | OmpF dependent | |
| Translation Rate | ||
| CFP | 0.167 /s | Based on effenciency RBS |
Cell Death
| Name | Value | Reference |
|---|---|---|
| Degradation | ||
| LuxR (protein) | 0.0010 /s | |
| LuxR (mRNA) | 0.00227 /s | [http://www.pubmedcentral.nih.gov/picrender.fcgi?artid=124983&blobtype=pdf link] |
| CcdB (protein) | 7.7E-5 /s | [http://www.ncbi.nlm.nih.gov/pubmed/8022284?dopt=abstract link] |
| CcdB (mRNA) | 0.00231 /s | [http://www.pubmedcentral.nih.gov/picrender.fcgi?artid=124983&blobtype=pdf link] |
| HSL | 1.02E-6/s | [http://aem.asm.org/cgi/content/abstract/71/3/1291 link] |
| Dissociation Rate | ||
| ka (HSL+LuxR) | 1E6 /s | chosen to be relatively (to the other rate constants) high and such that Kd equals 1E-6 |
| kd (HSL+LuxR) | 1 /s | |
| Dissociation Constant | ||
| HSL-LuxR | 1E-6 M/L | [http://jb.asm.org/cgi/content/full/189/11/4127?view=long&pmid=17400743 link] |
| Binding LuxR on LuxPromotor | 1E-9 M/L | [http://jb.asm.org/cgi/content/full/189/11/4127?view=long&pmid=17400743 link] |
| Transcription Rate | ||
| LuxR (constitutive promotor) | 0.025 mRNA/s | see sections on Constitutive Promotors & E. coli transcription Rates |
| CcdB (LuxR repressor) | k_1*K_m^n/(K_m^n + [LuxR]^n) = 0.025*1E-9/(1E-9^1+[LuxR]) |
Inverter
| Name | Value | Reference |
|---|---|---|
| Degradation | ||
| LuxI (protein) | 0.00231 /s | |
| LuxI (mRNA) | 0.0025 /s | [http://www.pubmedcentral.nih.gov/picrender.fcgi?artid=124983&blobtype=pdf link] |
| LuxI (complex) | 0.00125 /s | |
| LacI (protein) | 0.00231 /s | |
| LacI (gesloten mRNA) | 0.0046209812 /s | |
| LacI (open mRNA) | 0.0023104906 /s | |
| open mRNA LacI complex | 0.0023104906 /s | |
| HSL | 1.02E-6 /s | [http://aem.asm.org/cgi/content/abstract/71/3/1291 link] |
| Dissociation Rate | ||
| Transcription Rate | ||
| LacI | 0.0011 /s | |
| LuxI | 0.0025 /s | |
| K_m | 1.0E-10 /s | |
| Translation Rate | ||
| LuxI (RBS B0034) | 0.5 /s | |
| LacI (RBS ?) | 0.16667 /s | |
| Equilibrium Constant | ||
| closed and open mRNA LacI | 0.015 | |
| closed and complex mRNA LacI | 0.0212 /M | |
| Hill coëfficiënt | ||
| n | 2 | |
| Others | ||
| LuxI --> HSL | 0.16 | |
| LuxI (mRNA) + anti sense <-> complex LuxI | 4.22E14 | |
Memory
| Name | Value | Reference |
|---|---|---|
| Degradation | ||
| P2ogr (protein) | 0.002265 /s | |
| P2ogr (mRNA) | 0.002265 /s | [http://www.pubmedcentral.nih.gov/picrender.fcgi?artid=124983&blobtype=pdf link] |
| P22CII (protein) | 0.002311 /s | |
| P22CII (mRNA) | 0,0022651 /s | [http://www.pubmedcentral.nih.gov/picrender.fcgi?artid=124983&blobtype=pdf link] |
| antimRNA_luxI | 0.0045303/s | |
| Transcription rate | ||
| P2ogr | 0.0125 | |
| P22CII | 0.0125 | |
| AntimRNA | 0.0094 | |
| Dissociation Constant | ||
| P2-ogr | 4.2156 M | [http://www.jbc.org/cgi/content/abstract/258/17/10536?maxtoshow=&HITS=10&hits=10&RESULTFORMAT=&fulltext=P22+c2+repressor&andorexactfulltext=and&searchid=1&FIRSTINDEX=0&sortspec=relevance&volume=258&resourcetype=HWCIT link] |
| R0053-P22CII | 0.1099 M | |
| Hill coëfficiënt | ||
| n | 2 |
Filter
| Name | Value | Reference |
|---|---|---|
| Degradation | ||
| pT7_tag(protein) | 0.00155/s | |
| mRNA_RIBOKEY (RNA) | 0.00462 /s | [http://www.pubmedcentral.nih.gov/picrender.fcgi?artid=124983&blobtype=pdf link] |
| gesloten_mRNA_T7 (RNA) | 0.00462 /s | [http://www.pubmedcentral.nih.gov/picrender.fcgi?artid=124983&blobtype=pdf link] |
| open_mRNA_T7 (RNA) | 0.00231 / s | [http://www.pubmedcentral.nih.gov/picrender.fcgi?artid=124983&blobtype=pdf link] |
| open_mRNA_T7_complex (RNA) | 0.00231 /s | [http://www.pubmedcentral.nih.gov/picrender.fcgi?artid=124983&blobtype=pdf link] |
| Equilibrium Constant | ||
| closed and open mRNA | 0,015 | |
| closed and key complex mRNA | 0.0212 /M | |
| Transcription Rate | ||
| mRNA_RIBOKEY | Input dependent | |
| mRNA_T7 | 0,0011 mRNA/s |
Pulse Generator
Constant Lactonase production
Partial Model
Kinetic Constants
ETHZ list of parameters
mRNA decay
- [http://www.pubmedcentral.nih.gov/picrender.fcgi?artid=124983&blobtype=pdf Global analysis of mRNA decay and abundance in Escherichia coli at single-gene resolution using two-color fluorescent DNA microarrays]
T7 RNAP
- [http://www.jbc.org/cgi/reprint/267/4/2640 Initiation of Transcription by T7 RNA Polymerase at Its Natural Promoters]
- [http://pubs.acs.org/cgi-bin/article.cgi/bichaw/2002/41/i11/pdf/bi0158472.pdf Kinetic and Thermodynamic Basis of Promoter Strength: Multiple Steps of Transcription Initiation by T7 RNA Polymerase Are Modulated by the Promoter Sequence]
- [http://www.openwetware.org/wiki/IGEM:Tsinghua/2007/Projects/RAP data T7 team Tsinghua 2007]
- [http://www.jbc.org/cgi/reprint/281/47/35677 Transient State Kinetics of Transcription Elongation by T7 RNA Polymerase]
LacI - LuxI
- [http://www.tam.cornell.edu/tam/cms/manage/upload/Strogatz_20coupled_repressilators_PNAS.pdf Coupled repressilators]
- [http://www.nature.com/nature/journal/v403/n6767/full/403335a0.html A synthetic oscillatory network of transcriptional regulators]
LuxI, LuxR, mRNALuxI, mRNALuxR decay
- [http://bioinformatics.oxfordjournals.org/cgi/content/full/21/11/2722#E11 Noise-induced cooperative behavior in a multicell system]
- [http://www.pnas.org/content/93/18/9505 LuxI kinetics]
HSL stuff
- [http://aem.asm.org/cgi/content/abstract/71/3/1291 Rapid Acyl-Homoserine Lactone Quorum Signal Biodegradation in Diverse Soils] Fig 6: half-life 185 h ==> decay rate 1.02 * 10^-6 s^-1 ==> 0.00889 nM/h
- [http://www.sciencedirect.com/science?_ob=ArticleURL&_udi=B6WK7-457D0X9-9&_user=877992&_rdoc=1&_fmt=&_orig=search&_sort=d&view=c&_acct=C000047079&_version=1&_urlVersion=0&_userid=877992&md5=36d07f326b2c2e55c4de05a1ab426e09 Kinetics of the AHL Regulatory System in a Model Biofilm System]
- [http://www.pnas.org/content/96/8/4360.full HSL lactone synthesis kinetics by a LuxI-related enzyme]
- [http://www.jbc.org/cgi/content/full/279/14/13645#TBL2 Lactonase mediated HSL degradation (hydrolysis)]
OmpR, OmpF
- [http://www.pubmedcentral.nih.gov/picrender.fcgi?artid=543474&blobtype=pdf A simulation model of Escherichia coli osmoregulatory switch using E-CELL system] (Relevance: High), bekijk pg.11/13
- [http://www.jbc.org/cgi/reprint/281/25/17114 Transcription Regulation of ompF and ompC by a Single Transcription Factor, OmpR] (Relevance: Medium)
- [http://jb.oxfordjournals.org/cgi/reprint/111/6/707.pdf Transmembrane Signal Transduction and Osmoregulation in Escherichia coli: Functional Importance of the Transmembrane Regions of Membrane-Located Protein Kinase, EnvZ] (Relevance:?), geen toegang tenzij account
- [http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=209999 DNA-Binding Properties of the Transcription Activator (OmpR) for the Upstream Sequences of ompF in Escherichia coli Are Altered by envZ Mutations and Medium Osmolarity] (Relevance: Low)
- [http://jb.asm.org/cgi/reprint/176/5/1309 A Distant Upstream Site Involved in the Negative Regulation of the Escherichia coli ompF Gene] (Relevance: ?), voor repressor regulatie van OmpF door OmpR
- [http://jb.asm.org/cgi/reprint/176/16/5005.pdf micF Antisense RNA Has a Major Role in Osmoregulation of OmpF in Escherichia coli] (Relevance: Low), invloed 'Antisense RNA' op OmpF regulatie, hmm...
Psid met P2ogr promotor
- [http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=178405 Psid promotor with different activators]
Constitutive promoters
- Estimated transcription rate for J23105:[1]
- Scale other transcription rate with table in parts registry.
- Estimate the rate of transcription from a constitutive promotor family member.
E. coli transcription rates
[http://www.fasebj.org/cgi/content/summary/20/10/1721 Paper about the calculated transcription rates for every E.coli ORF]
[http://users.path.ox.ac.uk/~pcook/data/catalogs.html Tables with the calculated transcription rates for every E.coli ORF]
 "
"