Team:Minnesota/ModelingComparator
From 2008.igem.org
Emartin9808 (Talk | contribs) |
m (→SynBioSS Wiki) |
||
| (12 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
| - | |||
| - | |||
| - | |||
{| style="color:gold;background-color:#800000;" cellpadding="3" cellspacing="1" border="1" bordercolor="#fff" width="90%" align="center" | {| style="color:gold;background-color:#800000;" cellpadding="3" cellspacing="1" border="1" bordercolor="#fff" width="90%" align="center" | ||
!align="center"|[[Team:Minnesota|<font color="gold">Minnesota Home</font>]] | !align="center"|[[Team:Minnesota|<font color="gold">Minnesota Home</font>]] | ||
| Line 12: | Line 9: | ||
|} | |} | ||
| + | '''Computational Tools (all teams invited to use SynBioSS)''' | ||
| - | ===Mathematical Modeling for Team Comparator | + | Team Comparator is also developing two tools named the "SynBioSS Designer" and "SynBioSS Wiki". SynBioSS stands for '''Syn'''thetic '''Bio'''logy '''S'''oftware '''S'''uite. It is a software suite that can assist in synthetic biology applications. It is freely available at synbioss.sourceforge.net [http://synbioss.sourceforge.net]. As with any engineering problem, modeling the behavior of a putative synthetic biological system is a key step in the design process. To date, many of these simulations of synthetic systems have used coarse-grained models and deterministic kinetic equations. Our team, however, strives to use stochastic models spanning multiple time scales. One major bottleneck is that the detailed molecular-level reaction networks required for such simulations are time consuming to construct. Moreover, while many of the promoters, operators, repressors, etc. used in synthetic biology have been well studied, the necessary quantitative information is scattered throughout the literature. The SynBioSS Designer facilitates the construction of detailed and accurate models, while the SynBioSS Wiki addresses the challenges of data collection and curation. |
| + | |||
| + | |||
| + | == Computational Tools == | ||
| + | |||
| + | [[Image:SynBioSS_Designer.jpg|300px|right|thumb|A schematic representation of how the Designer translates a series of BioBricks into a kinetic model of the reaction network.]] | ||
| + | |||
| + | Both Designer and Wiki can be found via the SynBioSS [http://synbioss.sourceforge.net home page]. | ||
| + | |||
| + | === SynBioSS Designer === | ||
| + | |||
| + | SynBioSS Designer is an application for the automatic generation of sets of biomolecular reactions. This software allows a user to input the molecular parts involved in gene expression and regulation (e.g. promoters, transcription factors, ribosomes, etc.) The software then generates complete networks of reactions that represent transcription, translation, regulation, induction and degradation of those parts. To facilitate the creation of detailed kinetic models of synthetic gene networks composed of BioBricks, we have adapted SynBioSS Designer to automatically generate a kinetic model from a construct composed entirely of BioBricks. A NetCDF or SBML file is generated for simulations. | ||
| + | |||
| + | === SynBioSS Wiki === | ||
| + | |||
| + | [[Image:SynBioSS_Wiki.jpg|300px|right|thumb|SynBioSS Wiki's main page as of October 28th, 2008.]] | ||
| + | |||
| + | The inaccessibility of requisite kinetic data complicates the generation of detailed mechanistic models. We address this barrier by creating a web accessible database curated by users in a “Wiki” format. SynBioSS Wiki is a significant | ||
| + | extension of the open-source Mediawiki software. The Wiki stores reaction kinetic data in a formatted and searchable | ||
| + | scheme with references to the relevant literature. This framework allows for the input of reactions whose rates are described either by elementary first & second order rate equations or any arbitrarily complex rate equation defined using MathML (e.g. Hill type reactions). Reactions may be searched via participating molecules which may be proteins, DNA sequences, small molecules, etc. Once located, reactions of interest (along with their associated kinetic data) may be collected. The completed model may be exported in SBML format for additional editing or simulation. | ||
| + | |||
| + | It is also through the SynBioSS Wiki databases that SynBioSS Designer can access and proliferate kinetic information related to the simulation of BioBricks, thus extending the utility of the database for the benefit of the greater modeling community. To jumpstart the process, we have entered the known biomolecular interactions in the expression and regulation of well-studied operons, such as the lactose, the tetracycline and the arabinose operons. | ||
| + | |||
| + | ==Mathematical Modeling for Team Comparator== | ||
We will be modeling our system using our in-house software called Synthetic Biology Software Suite (SynBioSS is open source available http://synbioss.sourceforge.net/) and a network of reactions. | We will be modeling our system using our in-house software called Synthetic Biology Software Suite (SynBioSS is open source available http://synbioss.sourceforge.net/) and a network of reactions. | ||
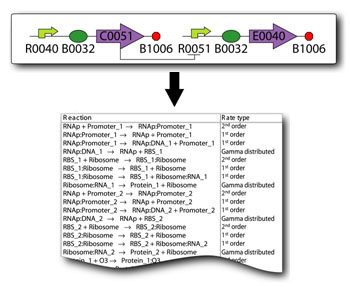
Our first try at the reaction network is shown below. | Our first try at the reaction network is shown below. | ||
{| | {| | ||
| - | |[[Media: | + | |[[Media:Comparator_Model_2-1.pdf|First Reaction Network]] |
| + | |} | ||
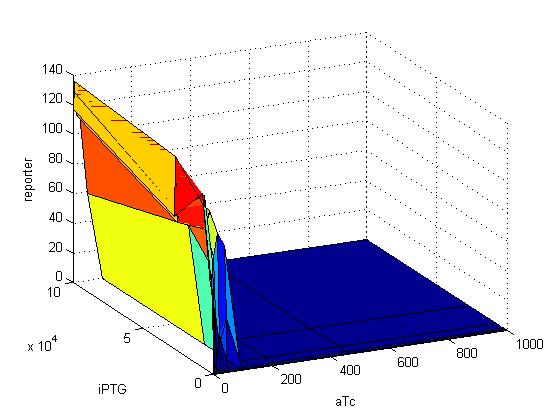
| + | {| | ||
| + | |[[Image:35gfp.jpg|500px|thumb]] | ||
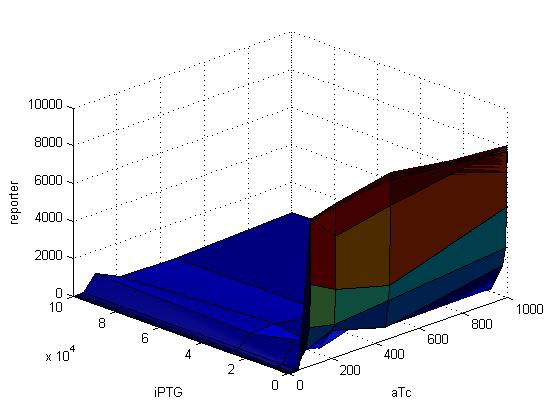
| + | |- | ||
| + | |[[Image:35rfp.jpg|500px|thumb]] | ||
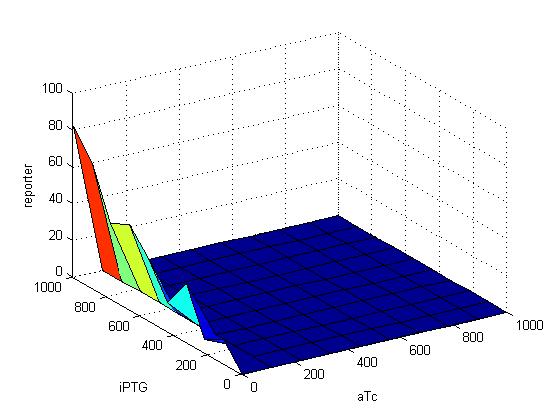
| + | |- | ||
| + | |[[Image:217GFP.jpg|500px|thumb]] | ||
| + | |- | ||
| + | |[[Image:217RFP.jpg|500px|thumb]] | ||
|} | |} | ||
Latest revision as of 22:49, 28 October 2008
| Minnesota Home | Team Comparator Home | The Team | The Project | Parts Submitted to the Registry | Modeling | Notebook |
|---|
Computational Tools (all teams invited to use SynBioSS)
Team Comparator is also developing two tools named the "SynBioSS Designer" and "SynBioSS Wiki". SynBioSS stands for Synthetic Biology Software Suite. It is a software suite that can assist in synthetic biology applications. It is freely available at synbioss.sourceforge.net [http://synbioss.sourceforge.net]. As with any engineering problem, modeling the behavior of a putative synthetic biological system is a key step in the design process. To date, many of these simulations of synthetic systems have used coarse-grained models and deterministic kinetic equations. Our team, however, strives to use stochastic models spanning multiple time scales. One major bottleneck is that the detailed molecular-level reaction networks required for such simulations are time consuming to construct. Moreover, while many of the promoters, operators, repressors, etc. used in synthetic biology have been well studied, the necessary quantitative information is scattered throughout the literature. The SynBioSS Designer facilitates the construction of detailed and accurate models, while the SynBioSS Wiki addresses the challenges of data collection and curation.
Contents |
Computational Tools
Both Designer and Wiki can be found via the SynBioSS [http://synbioss.sourceforge.net home page].
SynBioSS Designer
SynBioSS Designer is an application for the automatic generation of sets of biomolecular reactions. This software allows a user to input the molecular parts involved in gene expression and regulation (e.g. promoters, transcription factors, ribosomes, etc.) The software then generates complete networks of reactions that represent transcription, translation, regulation, induction and degradation of those parts. To facilitate the creation of detailed kinetic models of synthetic gene networks composed of BioBricks, we have adapted SynBioSS Designer to automatically generate a kinetic model from a construct composed entirely of BioBricks. A NetCDF or SBML file is generated for simulations.
SynBioSS Wiki
The inaccessibility of requisite kinetic data complicates the generation of detailed mechanistic models. We address this barrier by creating a web accessible database curated by users in a “Wiki” format. SynBioSS Wiki is a significant extension of the open-source Mediawiki software. The Wiki stores reaction kinetic data in a formatted and searchable scheme with references to the relevant literature. This framework allows for the input of reactions whose rates are described either by elementary first & second order rate equations or any arbitrarily complex rate equation defined using MathML (e.g. Hill type reactions). Reactions may be searched via participating molecules which may be proteins, DNA sequences, small molecules, etc. Once located, reactions of interest (along with their associated kinetic data) may be collected. The completed model may be exported in SBML format for additional editing or simulation.
It is also through the SynBioSS Wiki databases that SynBioSS Designer can access and proliferate kinetic information related to the simulation of BioBricks, thus extending the utility of the database for the benefit of the greater modeling community. To jumpstart the process, we have entered the known biomolecular interactions in the expression and regulation of well-studied operons, such as the lactose, the tetracycline and the arabinose operons.
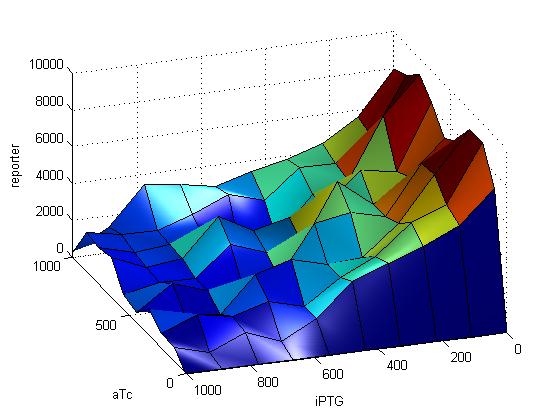
Mathematical Modeling for Team Comparator
We will be modeling our system using our in-house software called Synthetic Biology Software Suite (SynBioSS is open source available http://synbioss.sourceforge.net/) and a network of reactions. Our first try at the reaction network is shown below.
| First Reaction Network |
 "
"