Team:Paris/Modeling/Oscillations
From 2008.igem.org
(→Biochemical Assumptions) |
(→Resulting Equations) |
||
| Line 29: | Line 29: | ||
First, we introduce here what we assume to be in our model '''the involved chemical reactions'''. They are written in '''black''', regards to the equations that concerns both of the [[Team:Paris/Modeling/Oscillations#The_Circuit|alternatives]]. | First, we introduce here what we assume to be in our model '''the involved chemical reactions'''. They are written in '''black''', regards to the equations that concerns both of the [[Team:Paris/Modeling/Oscillations#The_Circuit|alternatives]]. | ||
| - | Then, the red and green equations are those implemented in our [[Team:Paris/Modeling/ | + | Then, the red and green equations are those implemented in our [[Team:Paris/Modeling/Codes|simulation programs]]. |
The '''blue''' ones correspond to the reactions of '''complexation''', and the '''red''' ones just below are the complexations functions that are their consequences if their stady-state are immediatly reached (see [[Team:Paris/Modeling/Oscillations#Biochemical_Assumptions|Biochemical Assumptions]]). | The '''blue''' ones correspond to the reactions of '''complexation''', and the '''red''' ones just below are the complexations functions that are their consequences if their stady-state are immediatly reached (see [[Team:Paris/Modeling/Oscillations#Biochemical_Assumptions|Biochemical Assumptions]]). | ||
Revision as of 17:21, 15 September 2008
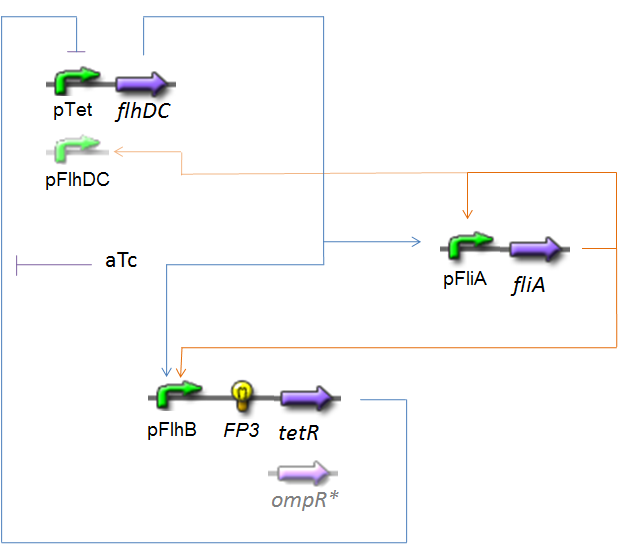
OscillationsThe CircuitWe just keep here the following circuit, constituing the Oscillations. The gene FP3 codes for the third reporter protein we will use in our final system. We have two alternatives for the promoter before flhDC : pTet or pFlhDC. These two alternatives are both studied in what follows.
Biochemical AssumptionsWe do not take into acount the phenomenon of translation : we consider the transduction as leading directly to the production of the protein (see however considerations on RBS). We assume that the expression rate of an inducible promoter is proportionnal to the number of created complexes promoter-inducer. In the same way, a repressible promoter has got a basal expression, and its expression is proportionnal to the number of free promoters. Moreover, even if generally the amount of a transcription factor is far superior than the amount of the corresponding promoter, because our complexation equation (see Team:Paris/Modeling/Programs), we have to take into acount that if a transcription factor binds to several promoters, those two complexations are in competition. In this way, we assume that all the steady-states of the complexations are reached at the same time, without taking into acount a possible kinetically predominant reaction : the singles complexation equations are put together in a consistent system. Endly, we suppose that the contribution of the two inducers FliA and FlhDC on PflhB are synthetised by a SUM logical gate. Resulting EquationsFirst, we introduce here what we assume to be in our model the involved chemical reactions. They are written in black, regards to the equations that concerns both of the alternatives. Then, the red and green equations are those implemented in our simulation programs. The blue ones correspond to the reactions of complexation, and the red ones just below are the complexations functions that are their consequences if their stady-state are immediatly reached (see Biochemical Assumptions). The green ones correspond to the reaction of production of proteins.
|
 "
"