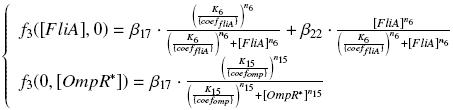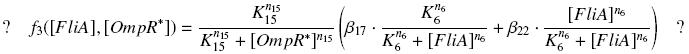Team:Paris/Modeling/f3
From 2008.igem.org
(Difference between revisions)
| (7 intermediate revisions not shown) | |||
| Line 2: | Line 2: | ||
{{Paris/Header|Method & Algorithm : ƒ3}} | {{Paris/Header|Method & Algorithm : ƒ3}} | ||
| + | <center> = act_''pFlhDC'' </center> | ||
| + | <br> | ||
[[Image:f3omp.jpg|thumb|Specific Plasmid Characterisation for ƒ3]] | [[Image:f3omp.jpg|thumb|Specific Plasmid Characterisation for ƒ3]] | ||
| - | + | According to the characterization plasmid (see right) and to our modeling, in the '''exponential phase of growth''', at the steady state, | |
| - | and | + | |
| - | + | we have ''' [''OmpR<sup>*</sup>'']<sub>''real''</sub> = {coef<sub>''ompR''</sub>} ƒ1([aTc]<sub>i</sub>) ''' | |
| - | and | + | and ''' [FliA]<sub>''real''</sub> = {coef<sub>FliA</sub>} ƒ2([arab]<sub>i</sub>) ''' |
| + | |||
| + | but we use ''' [aTc]<sub>i</sub> = Inv_ƒ1( [OmpR<sup>*</sup>] ) ''' | ||
| + | and ''' [arab]<sub>i</sub> = Inv_ƒ2( [FliA] ) ''' | ||
So, at steady-states, | So, at steady-states, | ||
| Line 15: | Line 19: | ||
[[Image:F3ompfinal.jpg|center]] | [[Image:F3ompfinal.jpg|center]] | ||
| - | + | we use this analytical expression to determine the parameters : | |
| - | + | ||
<div style="text-align: center"> | <div style="text-align: center"> | ||
| - | {{Paris/Toggle|Table|Team:Paris/Modeling/More_f3_Table}} | + | {{Paris/Toggle|Table of Values|Team:Paris/Modeling/More_f3_Table}} |
</div> | </div> | ||
| - | |||
<div style="text-align: center"> | <div style="text-align: center"> | ||
Latest revision as of 02:12, 30 October 2008
|
Method & Algorithm : ƒ3
According to the characterization plasmid (see right) and to our modeling, in the exponential phase of growth, at the steady state, we have [OmpR*]real = {coefompR} ƒ1([aTc]i) and [FliA]real = {coefFliA} ƒ2([arab]i) but we use [aTc]i = Inv_ƒ1( [OmpR*] ) and [arab]i = Inv_ƒ2( [FliA] ) So, at steady-states, we use this analytical expression to determine the parameters : ↓ Table of Values ↑
↓ Algorithm ↑
Then, if we have time, we want to verify the expected relation
<Back - to "Implementation" | |
 "
"