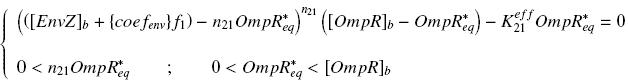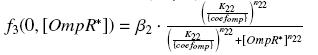Team:Paris/Modeling/f3bis
From 2008.igem.org
(Difference between revisions)
| Line 64: | Line 64: | ||
|comments | |comments | ||
|- | |- | ||
| - | | | + | |K<sub>21</sub><sup>eff</sup> |
| - | | | + | |dissociation constant OmpR_-_EnvZ <br> K<sub>21</sub><sup>eff</sup> |
|nM | |nM | ||
| | | | ||
| - | | | + | |the literature [[Team:Paris/Modeling/Bibliography|[?] ]] gives K<sub>21</sub> = |
|- | |- | ||
| - | |n<sub> | + | |n<sub>21</sub> |
| - | |complexation order | + | |complexation order OmpR_-_EnvZ <br> n<sub>21</sub> |
|no dimension | |no dimension | ||
| | | | ||
| - | | | + | |the literature [[Team:Paris/Modeling/Bibliography|[?] ]] gives n<sub>21</sub> = |
|- | |- | ||
| - | | | + | |{coef<sub>env</sub>} |
| - | | | + | |coefficient due to the difference of the RBS and degradation rate between EnvZ and GFP <br> ! not precised in the equations ! |
| - | + | |no dimension | |
| - | | | + | |
| | | | ||
| + | |! not precised in the equations ! watch out to write the corresponding simulating program | ||
|- | |- | ||
| - | | | + | |[EnvZ]<sub>b</sub> |
| - | | | + | |"basal" presence of EnvZ <br> [EnvZ]<sub>b</sub> |
|nM | |nM | ||
| | | | ||
| - | | | + | |the literature [[Team:Paris/Modeling/Bibliography|[?] ]] gives, under high osmolarity, [EnvZ]<sub>b</sub> = |
|- | |- | ||
| - | | | + | |[OmpR]<sub>b</sub> |
| - | | | + | |"basal" presence of OmpR <br> [OmpR]<sub>b</sub> |
| - | | | + | |nM |
| - | + | ||
| | | | ||
| + | |the literature [[Team:Paris/Modeling/Bibliography|[?] ]] gives, under high osmolarity, [OmpR]<sub>b</sub> = | ||
|} | |} | ||
Revision as of 11:17, 16 October 2008
(see the considerations on the use of EnvZ)We have [EnvZ]produced = {coefenv}expr(pTet) = {coefenv} ƒ1([aTc]i)
and [EnvZ]total = [EnvZ]b + [EnvZ]produced
and [FliA] = {coefFliA}expr(pBad) = {coefFliA} ƒ2([arab]i)
So, if we denote phosphorylated OmpR by OmpR*, we have
that we can then introduce in the previous expression (see ƒ3) :
| param | signification | unit | value | comments |
| [expr(pFlhDC)] | expression rate of pFlhDC with RBS E0032 | nM.min-1 | need for 20 mesures with well choosen values of [aTc]i | |
| γGFP | dilution-degradation rate of GFP(mut3b) | min-1 | 0.0198 | |
| [GFP] | GFP concentration at steady-state | nM | need for 20 | |
| (fluorescence) | value of the observed fluorescence | au | need for 20 | |
| conversion | conversion ratio between fluorescence and concentration | nM.au-1 | (1/79.429) |
| param | signification corresponding parameters in the equations | unit | value | comments |
| K21eff | dissociation constant OmpR_-_EnvZ K21eff | nM | the literature [?] gives K21 = | |
| n21 | complexation order OmpR_-_EnvZ n21 | no dimension | the literature [?] gives n21 = | |
| {coefenv} | coefficient due to the difference of the RBS and degradation rate between EnvZ and GFP ! not precised in the equations ! | no dimension | ! not precised in the equations ! watch out to write the corresponding simulating program | |
| [EnvZ]b | "basal" presence of EnvZ [EnvZ]b | nM | the literature [?] gives, under high osmolarity, [EnvZ]b = | |
| [OmpR]b | "basal" presence of OmpR [OmpR]b | nM | the literature [?] gives, under high osmolarity, [OmpR]b = |
 "
"