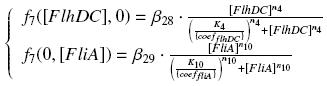|
|
| (6 intermediate revisions not shown) |
| Line 1: |
Line 1: |
| - | [[Image:f7DCA.png|thumb]]
| + | {{Paris/Menu}} |
| | | | |
| - | We have [FlhDC] = {coef<sub>flhDC</sub>}''expr(pTet)'' = {coef<sub>flhDC</sub>} ƒ1([aTc]<sub>i</sub>)
| + | {{Paris/Header|Method & Algorithm : ƒ7}} |
| | + | <center> = act_''pTet'' </center> |
| | + | <br> |
| | | | |
| - | and [FliA] = {coef<sub>FliA</sub>}''expr(pBad)'' = {coef<sub>FliA</sub>} ƒ2([arab]<sub>i</sub>)
| + | [[Image:f7DCA.png|thumb|Specific Plasmid Characterisation for ƒ7]] |
| | | | |
| - | So, at steady-states,
| + | According to the characterization plasmid (see right) and to our modeling, in the '''exponential phase of growth''', at the steady state, |
| | | | |
| - | [[Image:F7a.jpg|center]] | + | we have ''' [''FlhDC'']<sub>''real''</sub> = {coef<sub>''flhDC''</sub>} ƒ1([aTc]<sub>i</sub>) ''' |
| | + | and ''' [''FliA'']<sub>''real''</sub> = {coef<sub>''fliA''</sub>} ƒ2([arab]<sub>i</sub>) ''' |
| | | | |
| | + | but we use ''' [aTc]<sub>i</sub> = Inv_ƒ1( [''FlhDC''] ) ''' |
| | + | and ''' [arab]<sub>i</sub> = Inv_ƒ2( [''FliA''] ) ''' |
| | | | |
| - | {|border="1" style="text-align: center"
| + | So, at steady-states, |
| - | |param
| + | |
| - | |signification
| + | |
| - | |unit
| + | |
| - | |value
| + | |
| - | |comments
| + | |
| - | |-
| + | |
| - | |[expr(pFlhDC)]
| + | |
| - | |expression rate of <br> pFlhDC '''with RBS E0032'''
| + | |
| - | |nM.min<sup>-1</sup>
| + | |
| - | |
| + | |
| - | |need for 20 mesures with well choosen values of [aTc]<sub>i</sub> <br> and for 20 mesures with well choosen values of [arab]<sub>i</sub> <br> and 5x5 measures for the relation below?
| + | |
| - | |-
| + | |
| - | |γ<sub>GFP</sub>
| + | |
| - | |dilution-degradation rate <br> of GFP(mut3b)
| + | |
| - | |min<sup>-1</sup>
| + | |
| - | |0.0198
| + | |
| - | |
| + | |
| - | |-
| + | |
| - | |[GFP]
| + | |
| - | |GFP concentration at steady-state
| + | |
| - | |nM
| + | |
| - | |
| + | |
| - | |need for 20 + 20 measures <br> and 5x5 measures for the relation below?
| + | |
| - | |-
| + | |
| - | |(''fluorescence'')
| + | |
| - | |value of the observed fluorescence
| + | |
| - | |au
| + | |
| - | |
| + | |
| - | |need for 20 + 20 measures <br> and 5x5 measures for the relation below?
| + | |
| - | |-
| + | |
| - | |''conversion''
| + | |
| - | |conversion ratio between <br> fluorescence and concentration
| + | |
| - | |nM.au<sup>-1</sup>
| + | |
| - | |(1/79.429)
| + | |
| - | |
| + | |
| - | |}
| + | |
| | | | |
| - | <br><br>
| + | [[Image:F7a.jpg|center]] |
| | | | |
| - | {|border="1" style="text-align: center"
| + | we use this analytical expression to determine the parameters : |
| - | |param
| + | |
| - | |signification <br> corresponding parameters in the [[Team:Paris/Modeling/Oscillations#Resulting_Equations|equations]]
| + | |
| - | |unit
| + | |
| - | |value
| + | |
| - | |comments
| + | |
| - | |-
| + | |
| - | |β<sub>13</sub>
| + | |
| - | |production rate of FliA-pFlhDC '''with RBS E0032''' <br> β<sub>13</sub>
| + | |
| - | |nM.min<sup>-1</sup>
| + | |
| - | |
| + | |
| - | |
| + | |
| - | |-
| + | |
| - | |(K<sub>12</sub>/{coef<sub>fliA</sub>})
| + | |
| - | |activation constant of FliA-pFlhDC <br> K<sub>12</sub>
| + | |
| - | |nM
| + | |
| - | |
| + | |
| - | |
| + | |
| - | |-
| + | |
| - | |n<sub>12</sub>
| + | |
| - | |complexation order of FliA-pFlhDC <br> n<sub>12</sub>
| + | |
| - | |no dimension
| + | |
| - | |
| + | |
| - | |
| + | |
| - | |-
| + | |
| - | |β<sub>2</sub>
| + | |
| - | |production rate of OmpR-pFlhDC '''with RBS E0032''' <br> β<sub>2</sub>
| + | |
| - | |nM.min<sup>-1</sup>
| + | |
| - | |
| + | |
| - | |
| + | |
| - | |-
| + | |
| - | |(K<sub>22</sub>/{coef<sub>omp</sub>})
| + | |
| - | |activation constant of OmpR-pFlhDC <br> K<sub>22</sub>
| + | |
| - | |nM
| + | |
| - | |
| + | |
| - | |
| + | |
| - | |-
| + | |
| - | |n<sub>22</sub>
| + | |
| - | |complexation order of OmpR-pFlhDC <br> n<sub>22</sub>
| + | |
| - | |no dimension
| + | |
| - | |
| + | |
| - | |
| + | |
| - | |}
| + | |
| | | | |
| - | <br><br> | + | <div style="text-align: center"> |
| | + | {{Paris/Toggle|Table of Values|Team:Paris/Modeling/More_f7_Table}} |
| | + | </div> |
| | + | |
| | + | <div style="text-align: center"> |
| | + | {{Paris/Toggle|Algorithm|Team:Paris/Modeling/More_FP_Algo}} |
| | + | </div> |
| | | | |
| | Then, if we have time, we want to verify the expected relation | | Then, if we have time, we want to verify the expected relation |
| | | | |
| | [[Image:SumpFlgB.jpg|center]] | | [[Image:SumpFlgB.jpg|center]] |
| | + | |
| | + | <br> |
| | + | |
| | + | <center> |
| | + | [[Team:Paris/Modeling/Implementation| <Back - to "Implementation" ]]| <br> |
| | + | [[Team:Paris/Modeling/Protocol_Of_Characterization| <Back - to "Protocol Of Characterization" ]]| |
| | + | </center> |
|
Method & Algorithm : ƒ7
= act_pTet
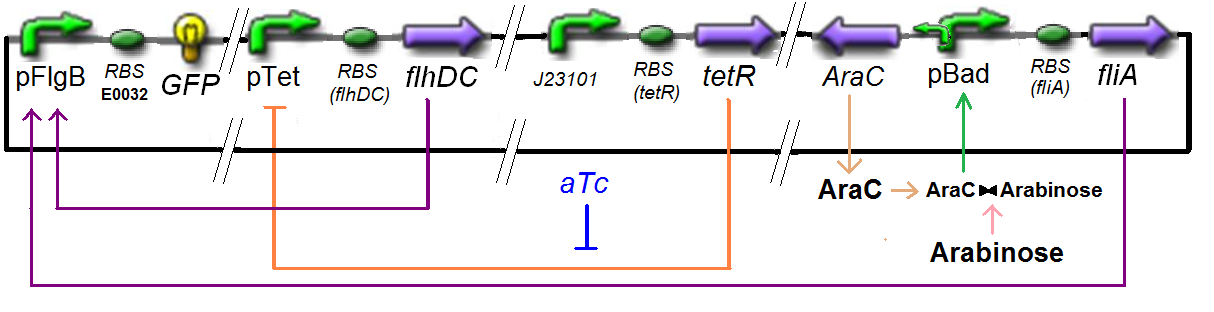 Specific Plasmid Characterisation for ƒ7 According to the characterization plasmid (see right) and to our modeling, in the exponential phase of growth, at the steady state,
we have [FlhDC]real = {coefflhDC} ƒ1([aTc]i)
and [FliA]real = {coeffliA} ƒ2([arab]i)
but we use [aTc]i = Inv_ƒ1( [FlhDC] )
and [arab]i = Inv_ƒ2( [FliA] )
So, at steady-states,
we use this analytical expression to determine the parameters :
↓ Table of Values ↑
| param
| signification
| unit
| value
| comments
|
| (fluorescence)
| value of the observed fluorescence
| au
|
| need for 20 mesures with well choosen values of [aTc]i
and for 20 mesures with well choosen values of [arab]i
and 5x5 measures for the relation below?
|
| conversion
| conversion ratio between
fluorescence and concentration
↓ gives ↓
| nM.au-1
| (1/79.429)
|
|
| [GFP]
| GFP concentration at steady-state
| nM
|
|
|
| γGFP
| dilution-degradation rate
of GFP(mut3b)
↓ gives ↓
| min-1
| 0.0198
| Time Cell Division : 35 min.
|
| ƒ7
| activity of
pFlgB with RBS E0032
| nM.min-1
|
|
|
| param
| signification
corresponding parameters in the equations
| unit
| value
| comments
|
| β28
| total transcription rate of
FlhDC><pFlgB with RBS E0032
β28
| nM.min-1
|
|
|
| (K4/{coeffliA})
| activation constant of FlhDC><pFlgB
K4
| nM
|
|
|
| n4
| complexation order of FlhDC><pFlgB
n4
| no dimension
|
|
|
| β29
| total transcription rate of
FlhDC><pFlgB with RBS E0032
β29
| nM.min-1
|
|
|
| (K10/{coefflhDC})
| activation constant of FlhDC><pFlgB
K10
| nM
|
|
|
| n10
| complexation order of FlhDC><pFlgB
n10
| no dimension
|
|
|
|
↓ Algorithm ↑
|
find_ƒP
function optimal_parameters = find_FP(X_data, Y_data, initial_parameters)
function output = act_pProm(parameters, X_data)
for k = 1:length(X_data)
output(k) = parameters(1)*hill(X_data(k), parameters(2), parameters(3));
end
end
options=optimset('LevenbergMarquardt','on','TolX',1e-10,'MaxFunEvals',1e10,'TolFun',1e-10,'MaxIter',1e4);
optimal_parameters = lsqcurvefit( @(parameters, X_data) act_pProm(parameters, X_data),...
initial_parameters, X_data, Y_data, options );
end
|
Then, if we have time, we want to verify the expected relation
<Back - to "Implementation" |
<Back - to "Protocol Of Characterization" |
|
 "
"