Team:Paris/Perspectives
From 2008.igem.org
m (Team:Paris/perspectives moved to Team:Paris/Perspectives) |
|||
| (31 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
{{Paris/Menu}} | {{Paris/Menu}} | ||
| + | {{Paris/Header|Perspectives}} | ||
| - | |||
| - | |||
| - | + | As for the flagella biosynthesis, our FIFO could be useful for many '''bottom-up assembled molecular machines''' that needs to be assembled in a precise order. | |
| + | Moreover, as we will show on an example, the FIFO could also be useful in many '''biosynthetic pathways''' subject to competitive alternative pathways and for wich intermediate products must be avoided. | ||
| + | We will develop below two examples: type III secretion injectisome and PHA biosynthesis pathway. | ||
| - | |||
| - | + | == Example 1: type III secretion injectisome == | |
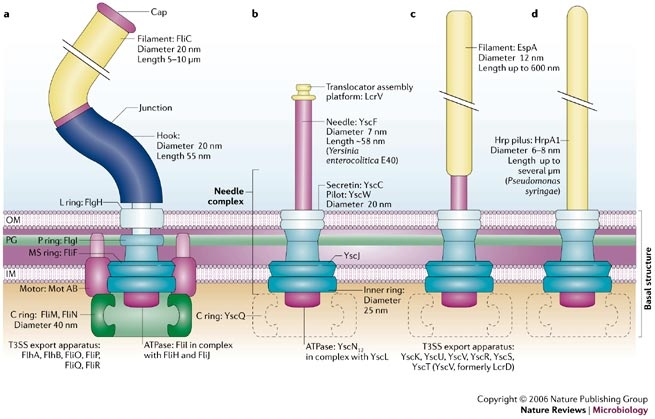
| - | + | The injectisome is a complex nanomachine that allows bacteria to deliver protein effector across eucaryotic cellular membranes. | |
| - | + | The injectisome consists of a basal structure, which resembles the basal structure of the flagellum, surmounted by either a needle, a needle and a filament or a long pilus. | |
| + | [[Image:injectisome.JPG]] | ||
| - | |||
| - | |||
| - | + | The injectisome is related to the flagellum genetically. Studies show that a core of eight proteins share significant similarity with components of the flagellum. From this studies, we can expect that the genes expression is ruled by a sequential FIFO order so we can express it with our bacteriO'clock system . | |
| + | Moreover, the injectisome will find a lot of applications for example ,bacteria that express this kinds of structure could : | ||
| + | * induce apoptosis of Eukaryote cells so we can developed by our system new kinds of in-vivo compatible anticancer therapy, | ||
| + | * be a new way to liberate substances produced by bacteria (see bioplastic application). | ||
| - | + | In conclusion, our BacteriO'Clock can be used in a lot of new applications like for instance, bottom-up molecular machines self-assembly or new kinds of optimized chemoreactor. | |
| - | + | == Example 2: metabolic engineering of polyhydroxyalkanoate biosynthesis pathways == | |
| + | Human overpopulation combined with the current lifestyle urges the rational, efficient, and sustainable use of natural resources to produce environmentally friendly plastic materials. | ||
| + | One illustrative example is polyhydroxyalkanoic acids (PHAs), whose production/degradation cycle reduces undesirable wastes and emissions. | ||
| + | The biosynthesis of this polymer is currently subject to intensive work. | ||
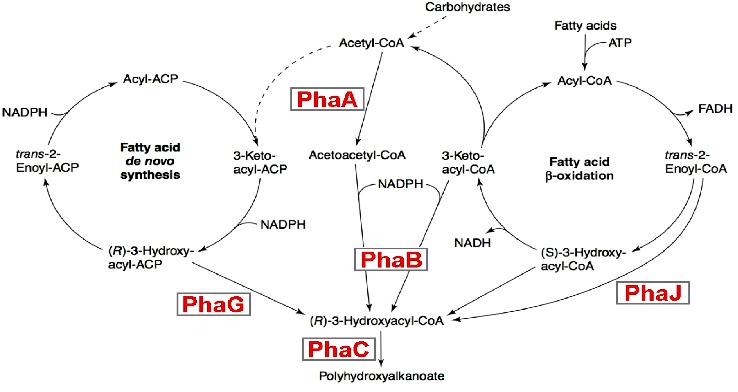
| + | It consists in expressing in appropriate quantities 3 enzymes PhaA ,PhaB and PhaC that sequentially process AcetoacetylcoA into its final product PHA. | ||
| + | This biosynthesis os subjected to 2 constraints : | ||
| + | * Intermediate products of this pathway are used in alternatives competing metabolic pathways, | ||
| + | * Only the final product is of interest,so that all intermediates products need to be transformed into PHA. | ||
| - | + | [[Image:PHA.jpg|center]] | |
| - | + | Two strategies are commonly used in bioengineering of metabolic pathways : | |
| + | * Sequential expression of enzymes involved in the pathways, | ||
| + | * Or constitutive expressions of all enzymes. | ||
| + | Because of the above mentioned limitations, none of these approaches are adapted here . | ||
| + | Using the sequential expression, intermediate products would accumulate and thus be consumed by competing pathways. | ||
| + | Using the constitutive expression a mixture of final and intermediate product would necessarily be obtained. | ||
| - | + | Our FIFO could be useful here. | |
| + | Indeed a FIFO expression pattern is intermediate between a purely sequential and a purely constitutive expression. | ||
| + | At some point all enzymes are presents ( no accumulation of intermediate products ) and during the last step only the last enzyme (PhC) is presents ( all intermediate products are consumed). | ||
| - | + | Moreover, the fact that our system oscillate could provide to the cell a metabolic recovering phase. | |
| - | |||
| - | + | == Bibliography == | |
| - | + | ||
| - | + | * The type III secretion injectisome Guy R.Cornelis Nature reviews | |
| + | * Process design for microbial plastic factories: metabolic engineering of polyhydroxyalkanoates | ||
| + | Ilana S Aldor and Jay D Keasling | ||
| + | Department of Chemical Engineering, 201 Gilman Hall, University of California, Berkeley, 94720-1462, USA | ||
| + | 23 September 2003 | ||
| - | + | * PHA synthase engineering toward superbiocatalysts for custom-made biopolymers. | |
| - | + | Nomura CT, Taguchi S. | |
| - | + | Department of Chemistry, State University of New York - College of Environmental Science and Forestry, 121 Jahn Laboratory, Syracuse, NY 13210, USA. | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
Latest revision as of 04:52, 30 October 2008
|
Perspectives
Example 1: type III secretion injectisomeThe injectisome is a complex nanomachine that allows bacteria to deliver protein effector across eucaryotic cellular membranes. The injectisome consists of a basal structure, which resembles the basal structure of the flagellum, surmounted by either a needle, a needle and a filament or a long pilus.
In conclusion, our BacteriO'Clock can be used in a lot of new applications like for instance, bottom-up molecular machines self-assembly or new kinds of optimized chemoreactor. Example 2: metabolic engineering of polyhydroxyalkanoate biosynthesis pathwaysHuman overpopulation combined with the current lifestyle urges the rational, efficient, and sustainable use of natural resources to produce environmentally friendly plastic materials. One illustrative example is polyhydroxyalkanoic acids (PHAs), whose production/degradation cycle reduces undesirable wastes and emissions. The biosynthesis of this polymer is currently subject to intensive work. It consists in expressing in appropriate quantities 3 enzymes PhaA ,PhaB and PhaC that sequentially process AcetoacetylcoA into its final product PHA. This biosynthesis os subjected to 2 constraints :
Two strategies are commonly used in bioengineering of metabolic pathways :
Because of the above mentioned limitations, none of these approaches are adapted here . Using the sequential expression, intermediate products would accumulate and thus be consumed by competing pathways. Using the constitutive expression a mixture of final and intermediate product would necessarily be obtained. Our FIFO could be useful here. Indeed a FIFO expression pattern is intermediate between a purely sequential and a purely constitutive expression. At some point all enzymes are presents ( no accumulation of intermediate products ) and during the last step only the last enzyme (PhC) is presents ( all intermediate products are consumed). Moreover, the fact that our system oscillate could provide to the cell a metabolic recovering phase.
Bibliography
Ilana S Aldor and Jay D Keasling Department of Chemical Engineering, 201 Gilman Hall, University of California, Berkeley, 94720-1462, USA 23 September 2003
Nomura CT, Taguchi S. Department of Chemistry, State University of New York - College of Environmental Science and Forestry, 121 Jahn Laboratory, Syracuse, NY 13210, USA. |
 "
"