|
← Yesterday ↓ Calendar ↑Tomorrow →
MiniPreps
- Use of Promega's protocol on all the clones cultivated on the 25th.
- Preparation of 50µl of minipreps in double.
| Name
| Biobrick
| Description
|
| MP100
| B0034
| Strongest RBS (Efficiency = 1)
|
| MP101
| J23101
| Strongest Constitutive promoter
|
| MP102
| J23109
| Weak Constitutive promoter
|
| MP103
| R0079
| promoter pLas (lasr & PAI regulated)
|
| MP104
| R0040
| tetR repressible promoter
|
| MP105
| S03154
| B0034(rbs) ->LasI
|
| MP106
| S03879
| B0034(rbs) -> TetR
|
| MP107
| C0079
| lasR activator with LVA Tag
|
| MP108
| C0179
| lasR activator
|
| MP109
| E0030
| YFP
|
| MP110
| E0040
| GFP
|
| MP111
| E1010
| mRFP
|
| MP116
| J23100
| Strong constitutive promoter in J61002
|
| MP117
| J23107
| Medium constitutive promoter in J61002
|
| MP118
| B0015
| Double terminator
|
| MP119
| I0500
| AraC pBAD
|
| MP120
| B0030
| Strong RBS (Efficiency = 0,6)
|
| MP121
| E0422
| ECFP (RBS+LVA+Term)
|
| MP122
| E0840
| gfp tri-part; strong rbs
|
|
Digestion
Digestion Mix
10µl of Miniprep (26 aug.)
12.5µl of water
2.5µl of Buffer N°2
0.25µl of BSA 100x
1µl of enzyme 1
1µl of enzyme 2
- Incubation 1h at 37°C with the first enzyme
- Add the second enzyme
- Incubation 1h at 37°C with the second enzyme
- Store on ice
- Revelation of the digestion by electrophoresis on agarose gel
Results of digestions : Electrophoresis
conditions :
- 10µl of ladder 1 kb (except for gel n°6 : 100 pb)
- 30µl of digestion added with 5µl of loading Dye 6x
- migration ~30min at 100W
- Gel 1, 2, 3, 4, 5 = 0.8%
- Gel 6 = 1,5%
gel1 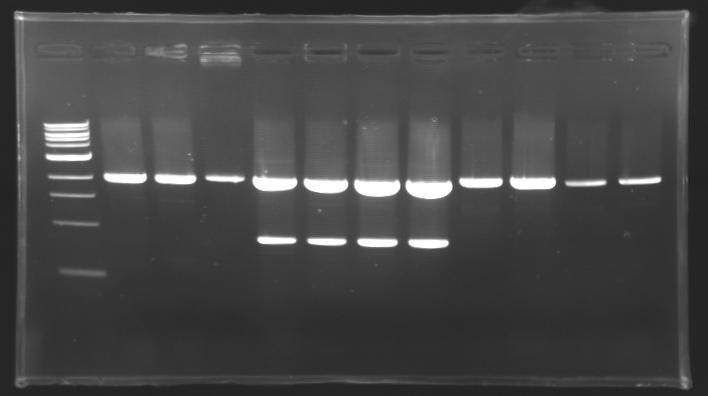 gel2
gel2 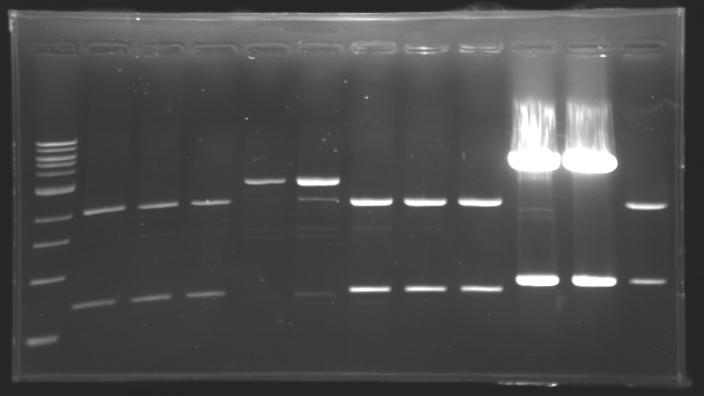 gel3
gel3 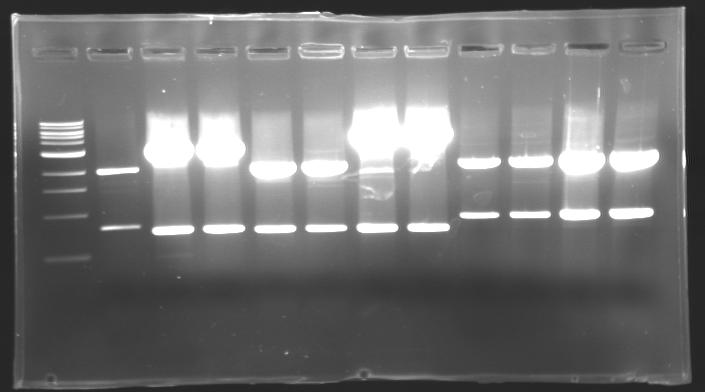 gel4
gel4 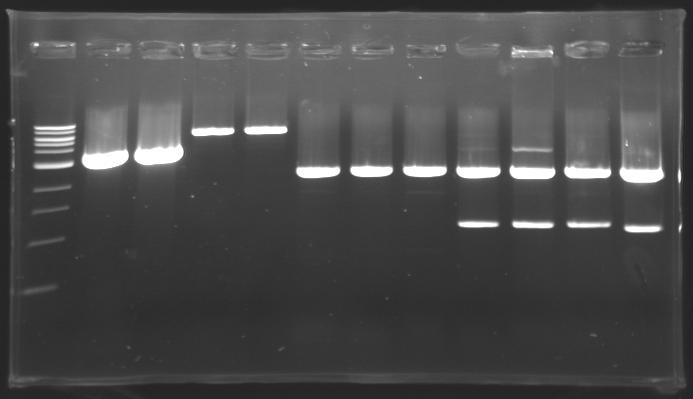
gel5 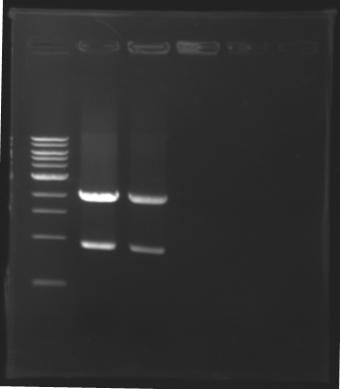 gel6
gel6 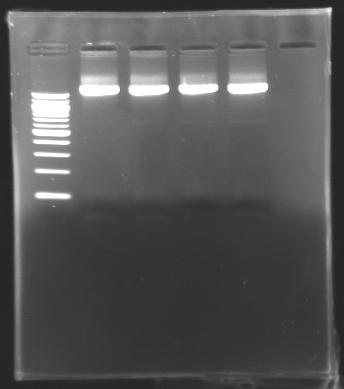
| Name
| BioBrick
| Tube N°
| Enz 1
| Enz 2
| Obs
| Exp Size Matrix
| Exp Size BB
| Mea Size Matrix
| Mea Size BB
| Gel
| Band
|
| D100
| B0034
| 1
| XbaI
| PstI
| BI
| 2057 pb
| 34 pb
| 1800 pb
| 40 pb
| 6
| 2
|
| 2
| 6
| 3
|
| D101
| B0034
| 3
| EcoRI
| XbaI
| FV
| 2076 pb
| 15 pb
| 2000 pb
| -
| 1
| 2
|
| D102
| B0034
| 4
| SpeI
| PstI
| BV
| 2077 pb
| 14 pb
| 2000 pb
| -
| 1
| 3
|
| 5
| 1
| 4
|
| D103
| J23101
| 1
| SpeI
| PstI
| BV
| 2100 pb
| 883 pb
| 2000 pb
| 700 pb
| 1
| 5
|
| 2
| 1
| 6
|
| D104
| J23109
| 1
| SpeI
| PstI
| BV
| 2100 pb
| 883 pb
| 2000 pb
| 700 pb
| 1
| 7
|
| 2
| 1
| 8
|
| D105
| R0079
| 1
| SpeI
| PstI
| BV
| 2222 pb
| 14 pb
| 2000 pb
| -
| 1
| 9
|
| 2
| 1
| 10
|
| D106
| R0040
| 1
| SpeI
| PstI
| BV
| 2119 pb
| 14 pb
| 2000 pb
| -
| 1
| 11
|
| 2
| 1
| 12
|
| D107
| S03154
| 1
| SpeI
| PstI
| BV
| 2750 pb
| 14 pb
| 3000 pb
| -
| 2
| 5
|
| D108
| S03154
| 2
| XbaI
| PstI
| BI
| 2057 pb
| 707 pb
| 2000 pb
| 700 pb
| 2
| 2
|
| 3
| 2
| 3
|
| D109
| S03154
| 4
| EcoRI
| SpeI
| FI
| 2056 pb
| 708 pb
| 2000 pb
| 700 pb
| 2
| 4
|
| D110
| S03879
| 1
| SpeI
| PstI
| BV
| 2768 pb
| 14 pb
| 2700 pb
| -
| 2
| 6
|
| D111
| S03879
| 2
| XbaI
| PstI
| BI
| 2057 pb
| 725 pb
| 2000 pb
| 700 pb
| 2
| 7
|
| 3
| 2
| 8
|
| D112
| S03879
| 4
| EcoRI
| SpeI
| FI
| 2756 pb
| 726 pb
| 3000 pb
| 700 pb
| 2
| 9
|
| D113
| C0079
| 1
| EcoRI
| SpeI
| FI
| 4402 pb
| 779 pb
| 5000 pb
| 800 pb
| 2
| 10
|
| D114
| C0079
| 2
| XbaI
| PstI
| BI
| 4003 pb
| 778 pb
| 5000 pb
| 800 pb
| 2
| 11
|
| D115
| C0179
| 1
| EcoRI
| SpeI
| FI
| 4402 pb
| 746 pb
| 2000 pb
| 800 pb
| 2
| 12
|
| D116
| C0179
| 2
| XbaI
| PstI
| BI
| 4403 pb
| 745 pb
| 2000 pb
| 800 pb
| 3
| 2
|
| D117
| E0030
| 1
| EcoRI
| SpeI
| FI
| 3166 pb
| 746 pb
| 3000 pb
| 700 pb
| 3
| 3
|
| D118
| E0030
| 2
| XbaI
| PstI
| BI
| 3167 pb
| 745 pb
| 3000 pb
| 700 pb
| 3
| 4
|
| D119
| E0040
| 1
| EcoRI
| SpeI
| FI
| 2056 pb
| 743 pb
| 2000 pb
| 700 pb
| 3
| 5
|
| D120
| E0040
| 2
| XbaI
| PstI
| BI
| 2057 pb
| 742 pb
| 2000 pb
| 700 pb
| 3
| 6
|
| D121
| E1010
| 1
| EcoRI
| SpeI
| FI
| 4402 pb
| 704 pb
| 5000 pb
| 700 pb
| 3
| 7
|
| D122
| E1010
| 2
| XbaI
| PstI
| BI
| 4403 pb
| 703 pb
| 5000 pb
| 700 pb
| 3
| 8
|
| D123
| J23100
| 1
| SpeI
| PstI
| BV
| 2100 pb
| 883 pb
| 2000 pb
| 900 pb
| 3
| 9
|
| 2
| 3
| 10
|
| D124
| J23107
| 1
| SpeI
| PstI
| BV
| 2100 pb
| 883 pb
| 2000 pb
| 900 pb
| 3
| 11
|
| 2
| 3
| 12
|
| D125
| B0015
| 1
| EcoRI
| XbaI
| BV
| 3303 pb
| 15 pb
| 3000 pb
| -
| 4
| 2
|
| 2
| 4
| 3
|
| D126
| I0500
| 1
| SpeI
| PstI
| FV
| 5621 pb
| 14 pb
| 6000 pb
| -
| 4
| 4
|
| 2
| 4
| 5
|
| D127
| B0030
| 1
| XbaI
| PstI
| BI
| 2057 pb
| 37 pb
| 2000 pb
| 50 pb
| 6
| 4
|
| 2
| 6
| 5
|
| D128
| B0030
| 3
| EcoRI
| XbaI
| FV
| 2079 pb
| 15 pb
| 2200 pb
| -
| 4
| 6
|
| D129
| B0030
| 4
| SpeI
| PstI
| BV
| 2080 pb
| 14 pb
| 2000 pb
| -
| 4
| 7
|
| 5
| 4
| 8
|
| D130
| E0422
| 1
| XbaI
| PstI
| BI
| 2057 pb
| 939 pb
| 2000 pb
| 1100 pb
| 4
| 9
|
| 2
| 4
| 10
|
| 3
| 4
| 11
|
| D131
| E0840
| 1
| XbaI
| PstI
| BI
| 2057 pb
| 900 pb
| 2000 pb
| 1000 pb
| 4
| 12
|
| 2
| 2000 pb
| 900 pb
| 5
| 2
|
| 3
| 5
| 3
|
==> conclusion : all the digestion have succeed.....GREAT !
- Cutting of the parts of interest, for all the digestion that have migrated on the gels
- Use of Promega's protocol for the extraction.
- Test of the succeed of the extraction by electrophoresis on 2µl of the parts extracted.
==> conclusion : we succeed to detect DNA in our samples this time.
|