Team:Paris/August 15
From 2008.igem.org
(Difference between revisions)
(→Analysis of yesterday PCR screening) |
(→Digestion) |
||
| (28 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
{{Paris/Calendar_Links|August 14|August 16}} | {{Paris/Calendar_Links|August 14|August 16}} | ||
| - | = | + | =Construction of OmpR*+RBS and EnvZ*+RBS= |
| - | I did some digestions(today), ligations( | + | I did some digestions (today), ligations (tommorow) and screening (the day after tommorow). |
I tried to build : | I tried to build : | ||
RBS (B0034) + OmpR* | RBS (B0034) + OmpR* | ||
| Line 38: | Line 38: | ||
* Incubate during about 3h at 37°C, then 20 minutes at 65°C (to inactivate the enzymes). Then 10°C overnight. | * Incubate during about 3h at 37°C, then 20 minutes at 65°C (to inactivate the enzymes). Then 10°C overnight. | ||
| - | = | + | =Creation of a registry of pFliL, pFlhDC, and ''FlhDC''= |
| + | ==Transformation of the ligations we did [[Team:Paris/August 14| yesterday]]== | ||
We transformed L 143, L 144 and the negative controls T1 and T2, using Invitrogen's TOP10 chemically competent cells [[Team:Paris/Notebook/Protocols#Transformation|standard protocol]]. | We transformed L 143, L 144 and the negative controls T1 and T2, using Invitrogen's TOP10 chemically competent cells [[Team:Paris/Notebook/Protocols#Transformation|standard protocol]]. | ||
| - | == | + | ==PCR amplification of flhDC and its promoter== |
==='''List of PCRs'''=== | ==='''List of PCRs'''=== | ||
| Line 184: | Line 185: | ||
We managed to amplify the promoter of flhDC | We managed to amplify the promoter of flhDC | ||
| - | == | + | ==Digestions== |
After having succeded in amplifying the promoter and the gene of flhDC, we decided to clone it into a plasmid. | After having succeded in amplifying the promoter and the gene of flhDC, we decided to clone it into a plasmid. | ||
The first step is the digestion. | The first step is the digestion. | ||
| Line 230: | Line 231: | ||
Then 10°C overnight. | Then 10°C overnight. | ||
| - | = | + | =Construction of pLas-TetR-GFP tripart & rbs-LasR-dble ter= |
| - | + | ==Measurement of concentration of minipreps== | |
[[Team:Paris/Notebook/Protocols#Concentration of the Miniprep|standard protocol]] | [[Team:Paris/Notebook/Protocols#Concentration of the Miniprep|standard protocol]] | ||
| Line 252: | Line 253: | ||
|} | |} | ||
| - | + | ==Digestion== | |
[[Team:Paris/Notebook/Protocols#Digestion|Protocol Digestion]] | [[Team:Paris/Notebook/Protocols#Digestion|Protocol Digestion]] | ||
| - | == | + | ==Ligation== |
[[Team:Paris/Notebook/Protocols#Ligation|Protocol Ligation]] | [[Team:Paris/Notebook/Protocols#Ligation|Protocol Ligation]] | ||
| Line 283: | Line 284: | ||
|} | |} | ||
| + | =PCR screening of cloning of E0240, flhD, flhC and pflhDC+flhDC= | ||
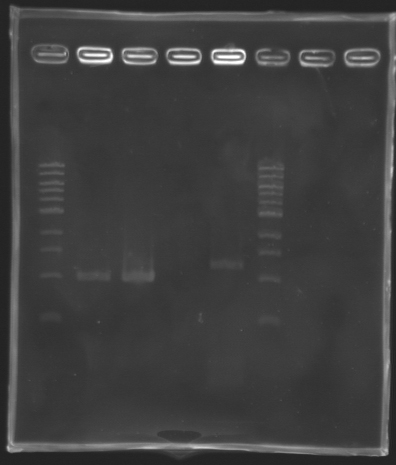
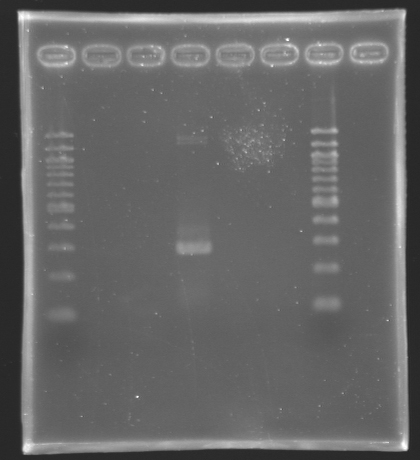
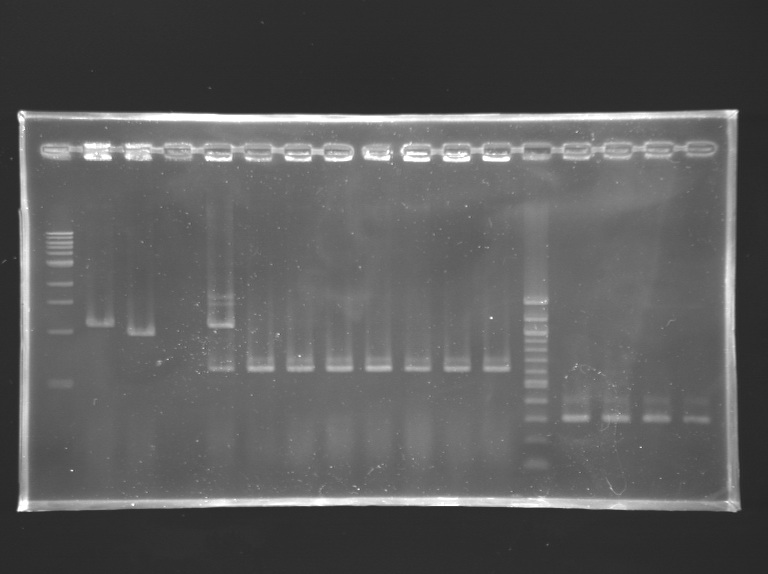
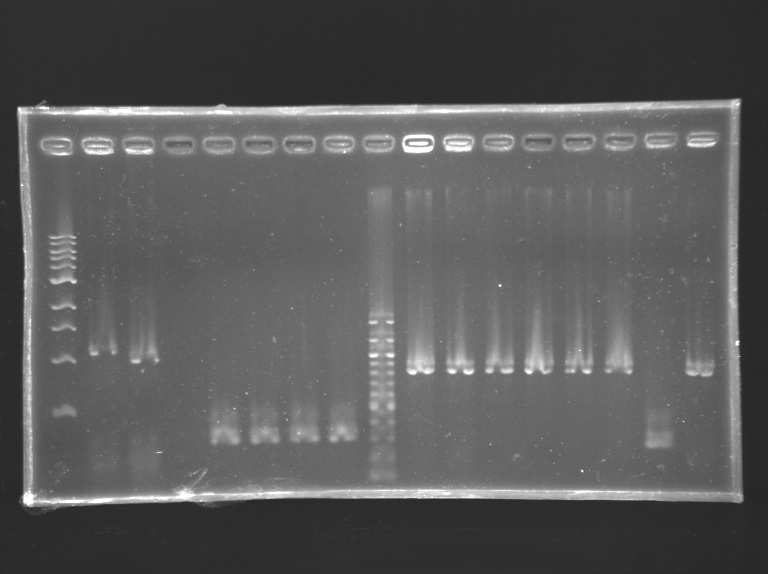
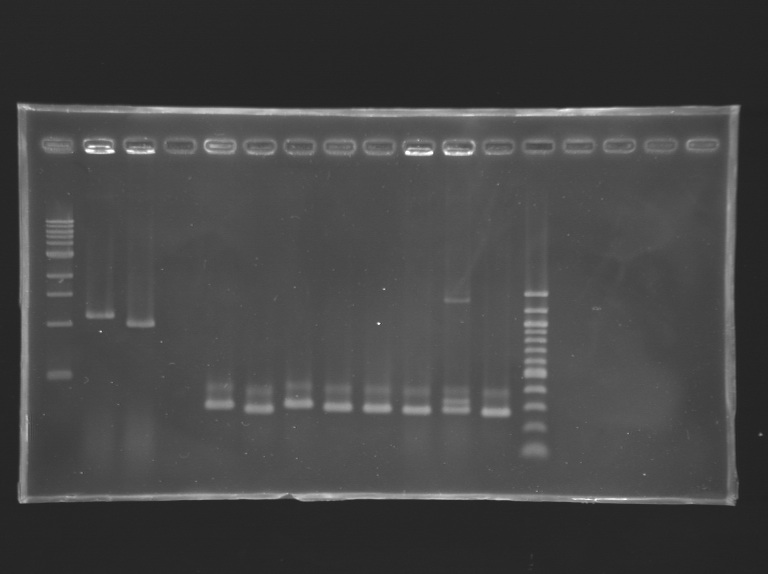
==Analysis of yesterday PCR screening== | ==Analysis of yesterday PCR screening== | ||
'''Electrophoresis''' | '''Electrophoresis''' | ||
| Line 288: | Line 290: | ||
*10 µL loaded | *10 µL loaded | ||
| + | Gel 1 [[Image:KR000163.jpg|200px|]] | ||
| + | Gel 2 [[Image:KR000162.jpg|200px|]] | ||
| + | Gel 3 [[Image:KR000164.jpg|200px|]] | ||
{| border="1" style="text-align: center" | {| border="1" style="text-align: center" | ||
|colspan="18"|'''Gel n°1''' | |colspan="18"|'''Gel n°1''' | ||
| Line 315: | Line 320: | ||
|positive control 2<br> pSB3K3 | |positive control 2<br> pSB3K3 | ||
|negative control | |negative control | ||
| + | |colspan="8"|'''E0240 in pSB3K3''' | ||
| + | |100 bp DNA ladder | ||
| + | |colspan="4"|'''FlhD in pSB1A2''' | ||
| + | |- | ||
| + | |'''clone''' | ||
| + | | | ||
| + | | | ||
| + | | | ||
| + | | | ||
|L139.1 | |L139.1 | ||
|L139.2 | |L139.2 | ||
| Line 323: | Line 337: | ||
|L139.7 | |L139.7 | ||
|L139.8 | |L139.8 | ||
| - | | | + | | |
|L140.1 | |L140.1 | ||
|L140.2 | |L140.2 | ||
| Line 334: | Line 348: | ||
|316 bp | |316 bp | ||
|0 kb | |0 kb | ||
| - | |colspan="8"|1192 bp | + | |colspan="8"|1192 bp |
| | | | ||
|colspan="4"|589 bp | |colspan="4"|589 bp | ||
| Line 343: | Line 357: | ||
|1 kb | |1 kb | ||
|0 kb | |0 kb | ||
| - | |1,1 kb<br>0,6 kb | + | |style="background: #cbff7B"|1,5 kb<br>'''1,1 kb'''<br>0,6 kb |
|0,6 kb | |0,6 kb | ||
|0,6 kb | |0,6 kb | ||
| Line 352: | Line 366: | ||
|0,6 kb | |0,6 kb | ||
| | | | ||
| - | |0,3 kb | + | |0,4 kb<br>0,3 kb |
| - | |0,3 kb | + | |0,4 kb<br>0,3 kb |
| - | |0,3 kb | + | |0,4 kb<br>0,3 kb |
| - | |0,3 kb | + | |0,4 kb<br>0,3 kb |
|} | |} | ||
| Line 385: | Line 399: | ||
|positive control 2<br> pSB3K3 | |positive control 2<br> pSB3K3 | ||
|negative control | |negative control | ||
| + | |colspan="4"|'''FlhD in pSB1A2''' | ||
| + | |100 bp DNA ladder | ||
| + | |colspan="8"|'''FlhC in pSB1A2''' | ||
| + | |- | ||
| + | |'''clone''' | ||
| + | | | ||
| + | | | ||
| + | | | ||
| + | | | ||
|L140.5 | |L140.5 | ||
|L140.6 | |L140.6 | ||
|L140.7 | |L140.7 | ||
|L140.8 | |L140.8 | ||
| - | | | + | | |
|L141.1 | |L141.1 | ||
|L141.2 | |L141.2 | ||
| Line 418: | Line 441: | ||
|0,3 kb | |0,3 kb | ||
| | | | ||
| - | | | + | |style="background: #cbff7B"|0,8 kb |
| - | | | + | |style="background: #cbff7B"|0,8 kb |
| - | | | + | |style="background: #cbff7B"|0,8 kb |
| - | | | + | |style="background: #cbff7B"|0,8 kb |
| - | | | + | |style="background: #cbff7B"|0,8 kb |
| - | | | + | |style="background: #cbff7B"|0,8 kb |
| - | | | + | |0,3 kb |
| - | | | + | |style="background: #cbff7B"|0,8 kb |
|} | |} | ||
| Line 455: | Line 478: | ||
|positive control 2<br> pSB3K3 | |positive control 2<br> pSB3K3 | ||
|negative control | |negative control | ||
| + | |colspan="8"|'''FlhDC+promotor in pSB1A2''' | ||
| + | |100 bp DNA ladder | ||
| + | | | ||
| + | | | ||
| + | | | ||
| + | | | ||
| + | |- | ||
| + | |'''clone''' | ||
| + | | | ||
| + | | | ||
| + | | | ||
| + | | | ||
|L142.1 | |L142.1 | ||
|L142.2 | |L142.2 | ||
| Line 463: | Line 498: | ||
|L142.7 | |L142.7 | ||
|L142.8 | |L142.8 | ||
| - | | | + | | |
| | | | ||
| | | | ||
| Line 483: | Line 518: | ||
|'''measured size''' | |'''measured size''' | ||
| | | | ||
| - | | | + | |1,1 kb |
| - | | | + | |1 kb |
|0 kb | |0 kb | ||
| - | | | + | |0,3 kb<br>0,4 kb |
| - | | | + | |0,3 kb<br>0,4 kb |
| - | | | + | |0,3 kb<br>0,4 kb |
| - | | | + | |0,3 kb<br>0,4 kb |
| - | | | + | |0,3 kb<br>0,4 kb |
| - | | | + | |0,3 kb<br>0,4 kb |
| - | | | + | |style="background: #cbff7B"|'''1,4 kb'''<br>0,4 kb<br>0,3 kb |
| - | | | + | |0,3 kb<br>0,4 kb |
| | | | ||
| | | | ||
| Line 500: | Line 535: | ||
| | | | ||
|} | |} | ||
| - | |||
| - | |||
| - | |||
| - | |||
=Starting the construction of the Promoter characterization plasmid= | =Starting the construction of the Promoter characterization plasmid= | ||
| - | |||
| - | + | ||
| + | ==Measurement of concentration of minipreps== | ||
[[Team:Paris/Notebook/Protocols#Concentration of the Miniprep|standard protocol]] | [[Team:Paris/Notebook/Protocols#Concentration of the Miniprep|standard protocol]] | ||
| Line 579: | Line 610: | ||
|} | |} | ||
| - | + | ==Digestion== | |
[[Team:Paris/Notebook/Protocols#Digestion|Protocol Digestion]] | [[Team:Paris/Notebook/Protocols#Digestion|Protocol Digestion]] | ||
{| border="1" style="text-align: center" | {| border="1" style="text-align: center" | ||
| + | |Name | ||
|Plasmid | |Plasmid | ||
|Description | |Description | ||
| Line 589: | Line 621: | ||
|Enzymes | |Enzymes | ||
|- | |- | ||
| + | | | ||
|MP3 | |MP3 | ||
|B0015 (double terminator B0010-B0012) - FV | |B0015 (double terminator B0010-B0012) - FV | ||
| Line 594: | Line 627: | ||
|EcoRI and XbaI | |EcoRI and XbaI | ||
|- | |- | ||
| + | |D164 | ||
|MP101 | |MP101 | ||
|promoter J23101 - BV | |promoter J23101 - BV | ||
| Line 599: | Line 633: | ||
|SpeI and PstI | |SpeI and PstI | ||
|- | |- | ||
| + | |D161 | ||
|MP104 | |MP104 | ||
|PTet (Tet promoter) - BV | |PTet (Tet promoter) - BV | ||
| Line 604: | Line 639: | ||
|SpeI and PstI | |SpeI and PstI | ||
|- | |- | ||
| + | |D162 | ||
|MP114 | |MP114 | ||
|TetR - FI | |TetR - FI | ||
| Line 609: | Line 645: | ||
|EcoRI and SpeI | |EcoRI and SpeI | ||
|- | |- | ||
| + | |D163 | ||
|MP143 | |MP143 | ||
|gfp generator - BI | |gfp generator - BI | ||
Latest revision as of 18:06, 4 September 2008
Construction of OmpR*+RBS and EnvZ*+RBSI did some digestions (today), ligations (tommorow) and screening (the day after tommorow). I tried to build : RBS (B0034) + OmpR* and RBS (B0034) + EnvZ* Protocol
Creation of a registry of pFliL, pFlhDC, and FlhDCTransformation of the ligations we did yesterdayWe transformed L 143, L 144 and the negative controls T1 and T2, using Invitrogen's TOP10 chemically competent cells standard protocol. PCR amplification of flhDC and its promoterList of PCRs
ProtocolWe followed the standard protocol of amplification in Two steps. PCR program used : PHUSION2 ResultsSettings Gel 1%
We managed to amplify flhDC ! Settings Gel 2%
We managed to amplify the promoter of flhDC DigestionsAfter having succeded in amplifying the promoter and the gene of flhDC, we decided to clone it into a plasmid. The first step is the digestion. Protocol
Then 10°C overnight. Construction of pLas-TetR-GFP tripart & rbs-LasR-dble terMeasurement of concentration of minipreps
DigestionLigation
PCR screening of cloning of E0240, flhD, flhC and pflhDC+flhDCAnalysis of yesterday PCR screeningElectrophoresis
Starting the construction of the Promoter characterization plasmidMeasurement of concentration of minipreps
Digestion
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
 "
"