Team:Paris/August 26
From 2008.igem.org
(→Cloning of EnvZ*) |
|||
| (32 intermediate revisions not shown) | |||
| Line 3: | Line 3: | ||
=Extraction of EnvZ* and OmpR* from ''E. coli'' genome= | =Extraction of EnvZ* and OmpR* from ''E. coli'' genome= | ||
==Name of the PCR== | ==Name of the PCR== | ||
| - | {|border="2" | + | {|style="text-align: center;" border="2" |
|Name | |Name | ||
|What's in ? | |What's in ? | ||
| Line 34: | Line 34: | ||
|O 139 | |O 139 | ||
|} | |} | ||
| + | |||
==Protocol== | ==Protocol== | ||
| Line 77: | Line 78: | ||
|- | |- | ||
|style="background: #cbff7B"|T 147 | |style="background: #cbff7B"|T 147 | ||
| - | | | + | |no sample |
|3 | |3 | ||
| - | | | + | |colspan="2"| |
| - | | | + | |
|- | |- | ||
|style="background: #cbff7B"|PCR 148 | |style="background: #cbff7B"|PCR 148 | ||
| Line 89: | Line 89: | ||
|- | |- | ||
|style="background: #cbff7B"|T 148 | |style="background: #cbff7B"|T 148 | ||
| - | | | + | |no sample |
|5 | |5 | ||
| - | | | + | |colspan="2"| |
| - | | | + | |
|} | |} | ||
Conclusion : All the PCR worked perfectly well ! | Conclusion : All the PCR worked perfectly well ! | ||
| Line 102: | Line 101: | ||
| - | =PCR= | + | =PCR Promoters and Genes FlhDC/FliA= |
| - | == | + | ==PCR Promoters FlhDC and Gene== |
| - | * pFlhDC (O111-F / O113-R) | + | * PCR 137 = pFlhDC (O111-F / O113-R) |
| - | * Gene FlhDC (O134-F / O131-R) | + | * PCR 141 = Gene FlhDC (O134-F / O131-R) |
| - | * pFlgB (O102-F / O103-R) | + | * PCR 125 = pFlgB (O102-F / O103-R) |
| - | * pFlhB (O108-F / O109-R) | + | * PCR 126 = pFlhB (O108-F / O109-R) |
---- | ---- | ||
| Line 135: | Line 134: | ||
Phusion=0,2µL<br> | Phusion=0,2µL<br> | ||
| - | == | + | ==PCR Promoters FliA== |
| + | [[Image:Autres_promoters.jpg|thumb|(49-60)PCR promoter FliA & Promoter+Gene]] | ||
* pFliA (rbs) (O145-F / O144-R) | * pFliA (rbs) (O145-F / O144-R) | ||
| Line 166: | Line 166: | ||
Phusion=0,5µL<br> | Phusion=0,5µL<br> | ||
| - | == | + | ==PCR mutagenesis FliA== |
| - | * PCRFliA1 (O143-F / O152-R) | + | * PCRFliA1 (O143-F / O152-R) |
| - | * PCRFliA2 (O153-F / O150-R) | + | * PCR 145 = PCRFliA1' (O148-F / O152-R) |
| + | * PCR 146 = PCRFliA2 (O153-F / O150-R) | ||
* PCRFliA3 (O148-F / O150-R) | * PCRFliA3 (O148-F / O150-R) | ||
---- | ---- | ||
| + | [[Image:PCRFliA1.jpg|thumb|(1-24)PCRFliA1]] | ||
| + | [[Image:PCRFliA2.jpg|thumb|(25-48)PCRFliA2]] | ||
| + | [[Image:PCRFliA1'.jpg|thumb|(61-72)PCRFliA1']] | ||
| + | [[Image:PCRFliA3.jpg|thumb|(73-84)PCRFliA3]] | ||
| + | |||
'''Program: PCRFliA1'''<br> | '''Program: PCRFliA1'''<br> | ||
| Line 180: | Line 186: | ||
98°C-10"<br> | 98°C-10"<br> | ||
Gradient 66°C +/-6°C - 25''<br> | Gradient 66°C +/-6°C - 25''<br> | ||
| + | 72°C-20"<br> | ||
| + | '''''Elongation :''''' <br> | ||
| + | 72°C-5'<br> | ||
| + | |||
| + | '''Program: PCRFliA1''''<br> | ||
| + | '''''Denaturation :''''' <br> | ||
| + | 98°C-5'<br> | ||
| + | '''''Cycling 1 (30X) :''''' <br> | ||
| + | 98°C-10"<br> | ||
72°C-20"<br> | 72°C-20"<br> | ||
'''''Elongation :''''' <br> | '''''Elongation :''''' <br> | ||
| Line 200: | Line 215: | ||
'''''Denaturation :''''' <br> | '''''Denaturation :''''' <br> | ||
98°C-30'<br> | 98°C-30'<br> | ||
| - | '''''Cycling 1 ( | + | '''''Cycling 1 (3X) :''''' <br> |
98°C-10"<br> | 98°C-10"<br> | ||
72°C-30"<br> | 72°C-30"<br> | ||
| Line 206: | Line 221: | ||
98°C-10"<br> | 98°C-10"<br> | ||
98°C->72°C low decreasing 1.0°C/s<br> | 98°C->72°C low decreasing 1.0°C/s<br> | ||
| + | 72°C-30"<br> | ||
| + | '''''Break - Add Oligo O148/O150'''''<br> | ||
| + | '''''Cycling 3 (20X) :'''''<br> | ||
| + | 98°C-10"<br> | ||
72°C-30"<br> | 72°C-30"<br> | ||
'''''Elongation :''''' <br> | '''''Elongation :''''' <br> | ||
| Line 222: | Line 241: | ||
|2-27 | |2-27 | ||
|197 bp | |197 bp | ||
| - | |style="background: #cbff7B"| ~ | + | |style="background: #cbff7B"| ~ 150 bp |
| + | |- | ||
| + | |PCRFliA1' | ||
| + | |Upstream part of FliA | ||
| + | |61-72 | ||
| + | |210 bp | ||
| + | |style="background: #cbff7B"| ~ 180 bp | ||
|- | |- | ||
|PCRFliA2 | |PCRFliA2 | ||
|Downstream part of FliA | |Downstream part of FliA | ||
| - | |28- | + | |28-53 |
| - | | | + | |670 bp |
| - | |style="background: #cbff7B"| | + | |style="background: #cbff7B"| ~ 650 bp |
| + | |- | ||
| + | |PCRFliA3 | ||
| + | |Mutated FliA | ||
| + | |73-84 | ||
| + | |800 bp | ||
| + | |style="background: #cbff7B"| ~ 800 bp | ||
|- | |- | ||
|PCR | |PCR | ||
|pFliA+rbs | |pFliA+rbs | ||
| - | | | + | |54-56 |
|325 bp | |325 bp | ||
| - | |style="background: #cbff7B"| | + | |style="background: #cbff7B"| ~ 350 bp |
|- | |- | ||
|PCR | |PCR | ||
| Line 240: | Line 271: | ||
|57-58 | |57-58 | ||
|310 bp | |310 bp | ||
| - | |style="background: #cbff7B"| | + | |style="background: #cbff7B"| ~ 320 bp |
|- | |- | ||
|PCR | |PCR | ||
| Line 246: | Line 277: | ||
|59-60 | |59-60 | ||
|1100 bp | |1100 bp | ||
| - | |style="background: #cbff7B"| | + | |style="background: #cbff7B"| ~ 1000 bp |
|} | |} | ||
| + | |||
=Cloning of EnvZ*= | =Cloning of EnvZ*= | ||
| Line 254: | Line 286: | ||
{|border="1" style="text-align:center" | {|border="1" style="text-align:center" | ||
| - | |||
|'''Digestion n°''' | |'''Digestion n°''' | ||
| + | |'''Name''' | ||
|'''[DNA] in µg/mL''' | |'''[DNA] in µg/mL''' | ||
|'''A260/A280''' | |'''A260/A280''' | ||
|- | |- | ||
| - | |||
|D159 | |D159 | ||
| + | |EnvZ* | ||
|10 µg/mL | |10 µg/mL | ||
|1.35 | |1.35 | ||
|- | |- | ||
| - | |||
|D116 | |D116 | ||
| + | |pSB1A2 | ||
|17 µg/mL | |17 µg/mL | ||
|1.02 | |1.02 | ||
| Line 319: | Line 351: | ||
*transformation of TOP10 competent cells with 5 µL of ligation product | *transformation of TOP10 competent cells with 5 µL of ligation product | ||
*spreading on LB plates + ampicilline and incubation overnight at 37°C | *spreading on LB plates + ampicilline and incubation overnight at 37°C | ||
| + | |||
| + | |||
='''Miniprep and stock glycerolof New Biobrick'''= | ='''Miniprep and stock glycerolof New Biobrick'''= | ||
| Line 324: | Line 358: | ||
*[[Team:Paris/Notebook/Protocols#Glycerol stocks| Protocol stocks]] | *[[Team:Paris/Notebook/Protocols#Glycerol stocks| Protocol stocks]] | ||
*[[Team:Paris/Notebook/Protocols#Minipreps (Kit_Qiagen)| Protocol miniprep]] | *[[Team:Paris/Notebook/Protocols#Minipreps (Kit_Qiagen)| Protocol miniprep]] | ||
| - | |||
| - | |||
{|border="1" style="text-align: center" | {|border="1" style="text-align: center" | ||
|'''Miniprep''' | |'''Miniprep''' | ||
|'''Strain''' | |'''Strain''' | ||
| + | |'''Antiobiotic''' | ||
|'''Name''' | |'''Name''' | ||
|'''Description''' | |'''Description''' | ||
|- | |- | ||
| - | | | + | |MP101.1 |
| - | | | + | |S109.1 |
| - | |rowspan="2"| | + | |rowspan="2"|Amp |
| - | |rowspan="2"| [[Image: | + | |rowspan="2"| J23101 |
| + | |rowspan="2"| [[Image:Part_icon_regulatory.png]]<br>constitutive promotor | ||
|- | |- | ||
| - | | | + | |MP101.2 |
| - | | | + | |S109.2 |
|- | |- | ||
| - | + | |MP101.1 | |
| - | | | + | |S109.1 |
| - | | | + | |rowspan="2"|Amp |
| - | |rowspan="2"| | + | |rowspan="2"| J23101 |
| - | |rowspan="2"| [[Image: | + | |rowspan="2"| [[Image:Part_icon_regulatory.png]]<br>constitutive promotor |
|- | |- | ||
| - | | | + | |MP101.2 |
| - | | | + | |S109.2 |
|- | |- | ||
| - | | | + | |MP104.1 |
| - | | | + | |S111.1 |
| - | |rowspan="2"| | + | |rowspan="2"|Amp |
| - | |rowspan="2"| [[Image: | + | |rowspan="2"| R0040 |
| + | |rowspan="2"| [[Image:Part_icon_regulatory.png]]<br>ptetR repressible | ||
|- | |- | ||
| - | | | + | |MP104.2 |
| - | | | + | |S111.2 |
|- | |- | ||
| - | | | + | |MP104.1 |
| - | | | + | |S111.1 |
| - | |rowspan="2"| | + | |rowspan="2"|Amp |
| - | |rowspan="2"| [[Image: | + | |rowspan="2"| R0040 |
| + | |rowspan="2"| [[Image:Part_icon_regulatory.png]]<br>ptetR repressible | ||
|- | |- | ||
| - | | | + | |MP104.2 |
| - | | | + | |S111.2 |
|- | |- | ||
| - | | | + | |MP105.1 |
| - | | | + | |S112.1 |
| - | |rowspan="2"| | + | |rowspan="2"|Amp |
| - | |rowspan="2"| [[Image:Icon_rbs.png]][[Image:Icon_reporter | + | |rowspan="2"| S03154 |
| + | |rowspan="2"| [[Image:Icon_rbs.png]][[Image:Icon_reporter.png]]<br>RBS-lasI | ||
|- | |- | ||
| - | | | + | |MP105.2 |
| - | | | + | |S112.2 |
| + | |- | ||
| + | |MP105.1 | ||
| + | |S112.1 | ||
| + | |rowspan="2"|Amp | ||
| + | |rowspan="2"| S03154 | ||
| + | |rowspan="2"| [[Image:Icon_rbs.png]][[Image:Icon_reporter.png]]<br>RBS-lasI | ||
| + | |- | ||
| + | |MP105.2 | ||
| + | |S112.2 | ||
| + | |- | ||
| + | |MP143 | ||
| + | |S157 | ||
| + | |Amp | ||
| + | |E0240 (in pSB1A2) | ||
| + | |[[Image:Icon_rbs.png]][[Image:Icon_reporter.png]][[Image:Part_icon_terminator.png]][[Image:Part_icon_terminator.png]]<br>RBS GFP dbl term | ||
| + | |- | ||
| + | |MP1151 | ||
| + | |S150 | ||
| + | |Amp | ||
| + | |L122 <br>D107 (BV) - D130 (BI) | ||
| + | |[[Image:Icon_rbs.png]][[Image:Icon_reporter.png]][[Image:Icon_rbs.png]][[Image:Icon_reporter.png]][[Image:Part_icon_terminator.png]][[Image:Part_icon_terminator.png]]<br>RBS-lasI - ECFP | ||
| + | |- | ||
| + | |MP157 | ||
| + | |S156 | ||
| + | |Kan | ||
| + | |L138 (E0240 in pSB3K3) | ||
| + | |[[Image:Icon_rbs.png]][[Image:Icon_reporter.png]][[Image:Part_icon_terminator.png]][[Image:Part_icon_terminator.png]]<br>gfp generator | ||
| + | |- | ||
| + | |MP158.1 | ||
| + | |S160.1 | ||
| + | |rowspan="3"|Amp | ||
| + | |rowspan="3"| L141 | ||
| + | |rowspan="3"| [[Image:Part_icon_regulatory.png]]<br>pFlhD | ||
| + | |- | ||
| + | |MP158.2 | ||
| + | |S160.2 | ||
| + | |- | ||
| + | |MP158.3 | ||
| + | |S160.3 | ||
| + | |- | ||
| + | |MP171 | ||
| + | |S171 | ||
| + | |Amp | ||
| + | |L167 <br>D181 (BV) - D184 (BI) | ||
| + | |[[Image:Icon_rbs.png]][[Image:Icon_reporter.png]][[Image:Part_icon_terminator.png]][[Image:Part_icon_terminator.png]][[Image:Part_icon_regulatory.png]]<br>gfp generator - pTet | ||
|} | |} | ||
| + | |||
| + | |||
='''Construction of pFlhB - mRFP Tripart (LVA+)'''= | ='''Construction of pFlhB - mRFP Tripart (LVA+)'''= | ||
| Line 401: | Line 485: | ||
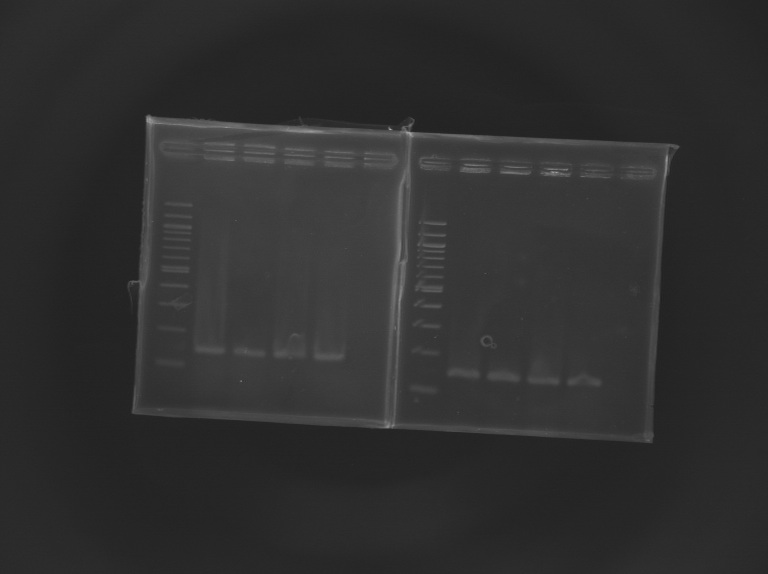
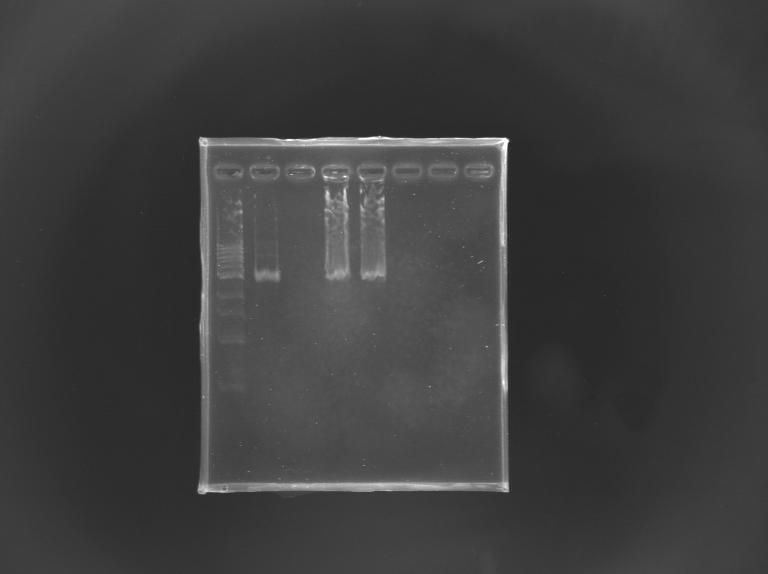
==='''Gel Verification'''=== | ==='''Gel Verification'''=== | ||
[[Team:Paris/Notebook/Protocols#Migration after extraction |Protocol]] | [[Team:Paris/Notebook/Protocols#Migration after extraction |Protocol]] | ||
| - | [[Image: | + | [[Image:KR000246.JPG |thumb |Gel Verification of D187 digestion]] |
{|border="1" style="text-align: center" | {|border="1" style="text-align: center" | ||
| Line 421: | Line 505: | ||
|'''Expected size (pb)''' | |'''Expected size (pb)''' | ||
| | | | ||
| + | |2 955 | ||
| | | | ||
| - | | | + | |2 940 |
| - | + | ||
| colspan="2"| | | colspan="2"| | ||
|- | |- | ||
|'''Measured size (pb)''' | |'''Measured size (pb)''' | ||
| | | | ||
| - | |style="background: #cbff7B"| | + | |style="background: #cbff7B"|2 900 |
| | | | ||
| - | |style="background: #cbff7B"| | + | |style="background: #cbff7B"|2 900 |
|colspan="2"| | |colspan="2"| | ||
|} | |} | ||
| Line 438: | Line 522: | ||
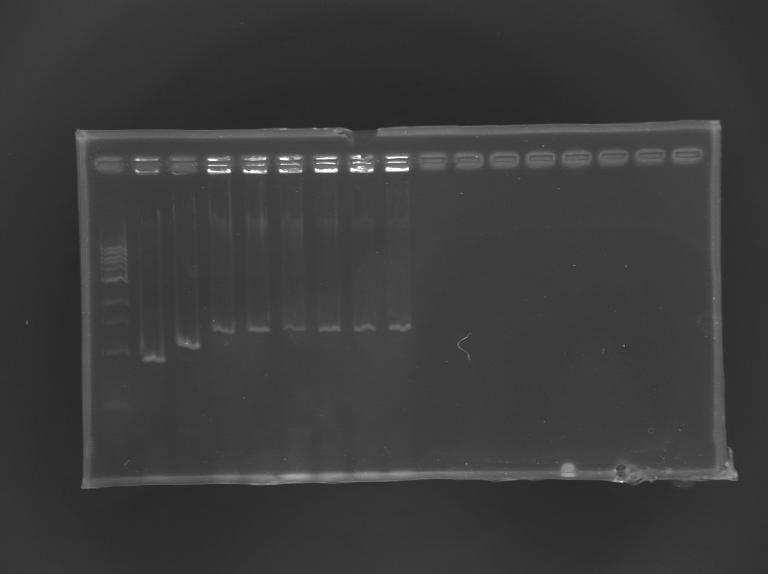
==Electrophoresis== | ==Electrophoresis== | ||
| + | [[Image:KR000245.JPG|thumb|Screening of L164]] | ||
{|border="1" style="text-align: center" | {|border="1" style="text-align: center" | ||
| Line 464: | Line 549: | ||
|'''expected size (pb)''' | |'''expected size (pb)''' | ||
| | | | ||
| - | | | + | |973 |
| - | | | + | |1176 |
| - | |colspan="6"|1 | + | |colspan="6"|1 426 |
|- | |- | ||
|'''measured size''' | |'''measured size''' | ||
| | | | ||
| - | | | + | |style="background: #cbff7B"|900 |
| - | |style="background: #cbff7B"| | + | |style="background: #cbff7B"|1 100 |
| - | |style="background: #cbff7B"| | + | |style="background: #cbff7B"|1 300 |
| - | |style="background: #cbff7B"| | + | |style="background: #cbff7B"|1 300 |
| - | |style="background: #cbff7B"| | + | |style="background: #cbff7B"|1 300 |
| - | |style="background: #cbff7B"| | + | |style="background: #cbff7B"|1 300 |
| - | |style="background: #cbff7B"| | + | |style="background: #cbff7B"|1 300 |
| - | |style="background: #cbff7B"| | + | |style="background: #cbff7B"|1 300 |
|} | |} | ||
| - | |||
==Minipreps and glycerol stock== | ==Minipreps and glycerol stock== | ||
| Line 487: | Line 571: | ||
|'''Glycerol Stock | |'''Glycerol Stock | ||
|'''Ligation''' | |'''Ligation''' | ||
| + | |'''Antibotic''' | ||
|'''Name''' | |'''Name''' | ||
|- | |- | ||
| - | | | + | |MP175.1 |
| - | | | + | |S177.1 |
| - | |L164 | + | |L164.1 |
| - | |FlgA- | + | |Amp |
| + | |[[Image:Part_icon_regulatory.png]][[Image:Part_icon_rbs.png]][[Image:icon_reporter.png]][[Image:Part_icon_terminator.png]][[Image:Part_icon_terminator.png]] | ||
| + | FlgA-GFP generator | ||
| + | |- | ||
| + | |MP175.2 | ||
| + | |S177.2 | ||
| + | |L164.2 | ||
| + | |Amp | ||
| + | |[[Image:Part_icon_regulatory.png]][[Image:Part_icon_rbs.png]][[Image:icon_reporter.png]][[Image:Part_icon_terminator.png]][[Image:Part_icon_terminator.png]] | ||
| + | FlgA-GFP generator | ||
| + | |- | ||
| + | |MP175.4 | ||
| + | |S177.4 | ||
| + | |L164.4 | ||
| + | |Amp | ||
| + | |[[Image:Part_icon_regulatory.png]][[Image:Part_icon_rbs.png]][[Image:icon_reporter.png]][[Image:Part_icon_terminator.png]][[Image:Part_icon_terminator.png]] | ||
| + | FlgA-GFP generator | ||
| + | |} | ||
| + | |||
| + | |||
| + | ='''Construction for Synchronisation'''= | ||
| + | =='''Results of the transformation we did [[Team:Paris/August 24 |yesterday]]'''== | ||
| + | |||
| + | {|border="1" style="text-align: center" | ||
| + | |'''Ligation name''' | ||
| + | |'''Description''' | ||
| + | |'''Antibio''' | ||
| + | |'''Number Colonies observed''' | ||
| + | |'''Fluorescence''' | ||
| + | |'''Comments''' | ||
| + | |- | ||
| + | |colspan="6" |Ligations | ||
| + | |- | ||
| + | |L159 | ||
| + | |D109(FI) - D125(FV)<br>rbs-lasI - double terminator | ||
| + | |Kan | ||
| + | | - | ||
| + | | - | ||
| + | | to do again | ||
| + | |- | ||
| + | |L162 | ||
| + | |D107(BV) - D163(BI)<br>rbs-lasI - gfp generator | ||
| + | |Amp | ||
| + | | - | ||
| + | | - | ||
| + | | to do again | ||
| + | |- | ||
| + | |L162 | ||
| + | |D110(BV) - D163(BI)<br>rbs-TetR - gfp generator | ||
| + | |Amp | ||
| + | | - | ||
| + | | - | ||
| + | | to do again | ||
| + | |- | ||
| + | |colspan="6" |Controls | ||
| + | |- | ||
| + | |TL159 | ||
| + | |D125(FV) | ||
| + | |Kan | ||
| + | | - | ||
| + | | - | ||
| + | |to do again | ||
| + | |- | ||
| + | |TL162 | ||
| + | |D107(BV) | ||
| + | |Amp | ||
| + | | - | ||
| + | | - | ||
| + | |to do again | ||
| + | |- | ||
| + | |TL163 | ||
| + | |D110(BV) | ||
| + | |AMp | ||
| + | | - | ||
| + | | - | ||
| + | |to do again | ||
| + | |- | ||
| + | |Positive Control | ||
| + | |pUC19 | ||
| + | |Amp | ||
| + | | | ||
| + | |No | ||
| + | |OK | ||
|} | |} | ||
Latest revision as of 17:59, 11 September 2008
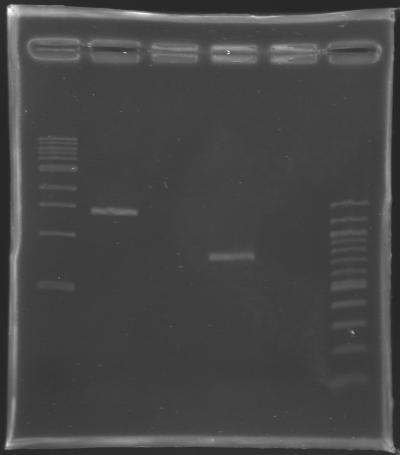
Extraction of EnvZ* and OmpR* from E. coli genomeName of the PCR
ProtocolProtocol
PCR Programme
ResutsElectrophoresis settings
Results of the electrophoresis
Conclusion : All the PCR worked perfectly well ! Cleaning of the PCR productsThe cleaned PCR products are stored in the freezer overnight.
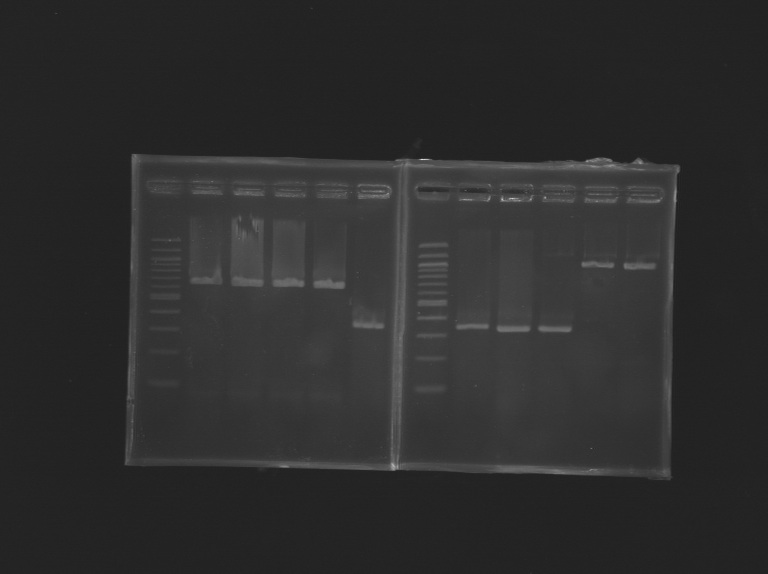
PCR Promoters and Genes FlhDC/FliAPCR Promoters FlhDC and Gene
Program: Gradient Vf=20µL PCR Promoters FliA
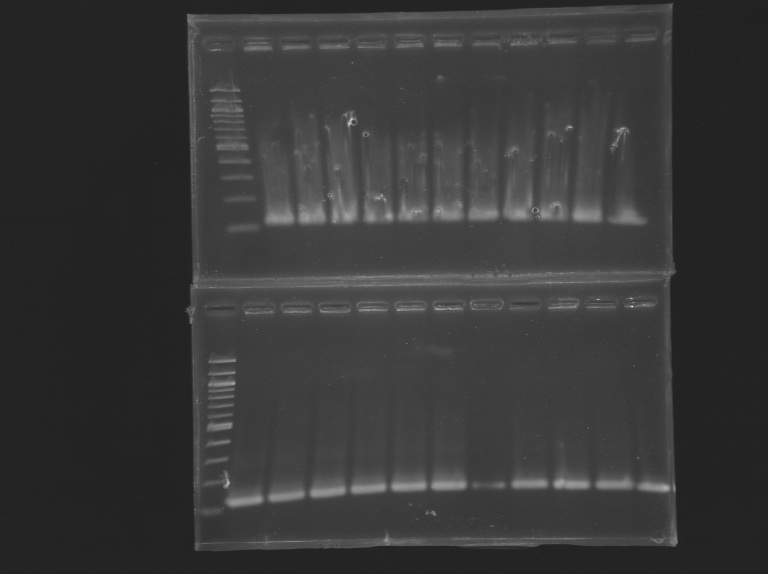
Program: promoter Vf=50µL PCR mutagenesis FliA
Program: PCRFliA1' Program: PCRFliA2 Program: PCRFliA3
Cloning of EnvZ*Concentration measurement by Biophotometer
Ligation
Miniprep and stock glycerolof New Biobrick
Construction of pFlhB - mRFP Tripart (LVA+)Aim : Construction of "pFlhB-RBS-mRFP-dbl ter" (pFlhB-I732078) DigestionDigestion
Gel Verification
Screening of the cloning of pFlgA-GFP GeneratorElectrophoresis
Minipreps and glycerol stock
Construction for SynchronisationResults of the transformation we did yesterday
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
 "
"