Alberta NINT/9 June 2008
From 2008.igem.org
(Difference between revisions)
m |
|||
| Line 1: | Line 1: | ||
[[Team:Alberta_NINT/Notebook | < Back to notebook]] | [[Team:Alberta_NINT/Notebook | < Back to notebook]] | ||
| - | [[Alberta_NINT/6_June_2008 | < Previous entry]] | + | [[Alberta_NINT/6_June_2008 | < Previous entry]] | [[Alberta_NINT/10_June_2008 | Next entry >]] |
lab work (SD): | lab work (SD): | ||
| Line 33: | Line 33: | ||
K102005 = 2116 + 362 | K102005 = 2116 + 362 | ||
[[Image:annotated1.jpg]] | [[Image:annotated1.jpg]] | ||
| - | |||
| - | |||
Latest revision as of 22:43, 11 October 2008
< Previous entry | Next entry >
lab work (SD):
K102008 and K102008 negative control plates again had >100 colonies each.
Verified sequencing for K102002.
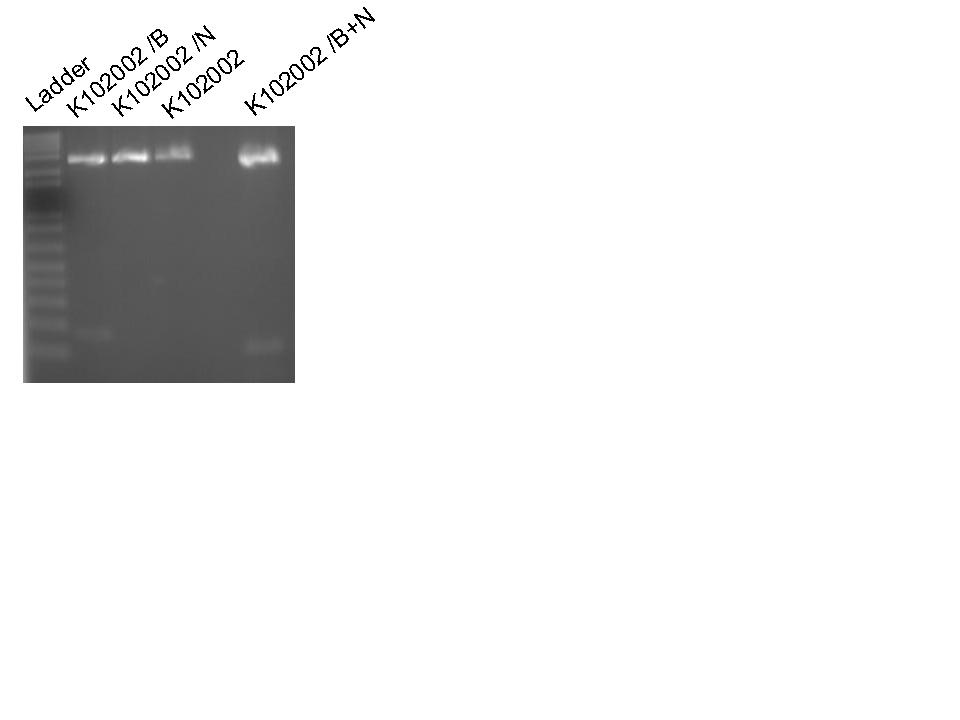
Digested K102002 with BamHI, with NcoI, and with BamHI+NcoI to test the digestion.
Ran DNA from digest on 2% agarose gel.
Gel reveals that digest of K102002 with BamHI formed two fragments. Conclusion: K102002 contains
two BamHI restriction sites. A closer examination of the sequences indicates BBa_E0433 (LacZ alpha)
contains a BamHI restriction site. A new version of LacZ alpha is required.
BBa_I732018 is LacZ alpha but lacks the BamHI restriction site. The DNA for this part is isolated
from the iGEM BioBricks parts registry book according to the standard procedure.
lab work (JD):
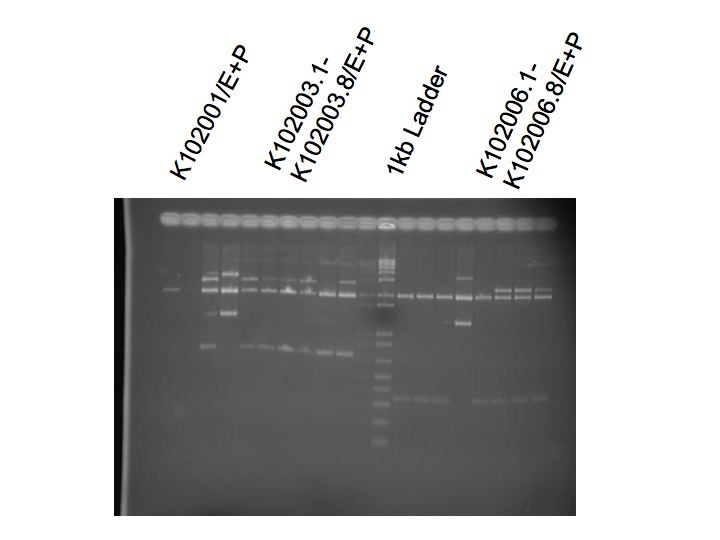
Wayne created O/N colonies of K102003 and K102005, 8 from each
Mini-prep of all colonies
Digest K102001/K102003/K102005 with EcoRI and PstI
Ran a gel
lab work (WM):
Performed minipreps on K102034, 2035, 2036 O/Ns
Sequenced with VR
K102001 = 2116+180
K102003 = 2116+733
K102005 = 2116 + 362
 "
"