Team:BCCS-Bristol/Calendar-Notebook/23 July 2008
From 2008.igem.org
Rodgerread (Talk | contribs) |
Tgorochowski (Talk | contribs) |
||
| (10 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
| - | <html><link rel="stylesheet" href="http:// | + | <html><link rel="stylesheet" href="http://homepage.mac.com/tgorochowski/iGEM/bccs-igem.css" type="text/css"></html> |
__NOTOC__ | __NOTOC__ | ||
<div class="bccsNavBar"> | <div class="bccsNavBar"> | ||
| Line 15: | Line 15: | ||
</div> | </div> | ||
| + | ==Swimming agar assay== | ||
| - | Results for | + | The experiment from [[Team:BCCS-Bristol/Calendar-Notebook/21 July 2008 | 21 July 2008]] was repeated. The aspartate solution was added eigth hours before the inoculation. The Results for showed that both strains were actually repelled by aspartate, possibly because the concentration was too high. Aspartate is the well on the right, and the control (water) is on the left. |
| + | {| align="center" | ||
| + | !align="center"|[[Image:BCCS-WetLabMC1000_25C_1.JPG | 200px]] | ||
| + | !align="center"|[[Image:BCCS-WetLabMC1000_30C_3.JPG | 200px]] | ||
| + | !align="center"|[[Image:BCCS-WetLabMG1655_25C_3.JPG | 200px]] | ||
| + | !align="center"|[[Image:BCCS-WetLabMG1655_30C_5.JPG | 200px]] | ||
| + | |} | ||
| + | {| align="center" | ||
| + | !align="center" width="10%"|MC1000 25<sup>o</sup>C | ||
| + | !align="center" width="10%"|MC1000 30<sup>o</sup>C | ||
| + | !align="center" width="10%"|MG1655 25<sup>o</sup>C | ||
| + | !align="center" width="10%"|MG1655 30<sup>o</sup>C | ||
| + | |} | ||
| + | |||
| + | ==BioBrick Transformation== | ||
| - | The cells transformed with | + | The cells transformed with BioBrick were grown on ampicillin plates as if transformed are able to do so, however there was no growth of bacteria except when pUC19 was used. This highlights some problems with the DNA and not the actual transformation. The experiment was repeated with larger amounts of TE buffer around spot and with a different BioBrick. Results see weekly summary. |
| - | + | ==Bead experiment 1== | |
| - | The bead we had focused on had gone!! lots of beads | + | The bead we had focused on overnight (see [[Team:BCCS-Bristol/Calendar-Notebook/22 July 2008 | 22 July 2008]]) had gone!! There were observed lots of beads clumping together, areas of dead/non-motile bacteria and also filamentous bacteria (possible conaminant). There were also areas of still motile bacteria. |
| - | bacteria had travelled 40% of way across bridge, | + | The bacteria had travelled 40 % of way across bridge, thus a smaller bridge was suggested. |
Latest revision as of 09:56, 14 September 2008
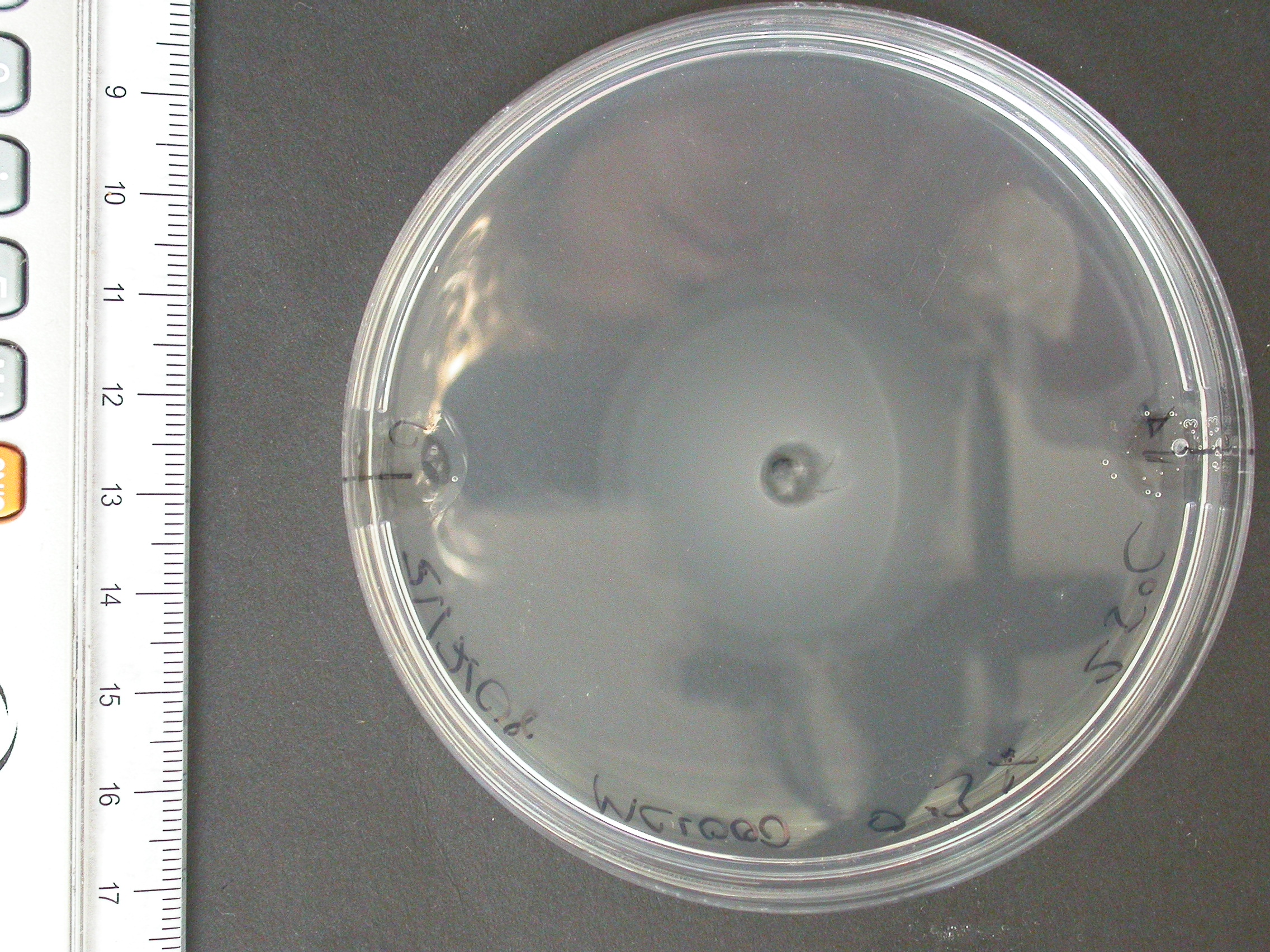
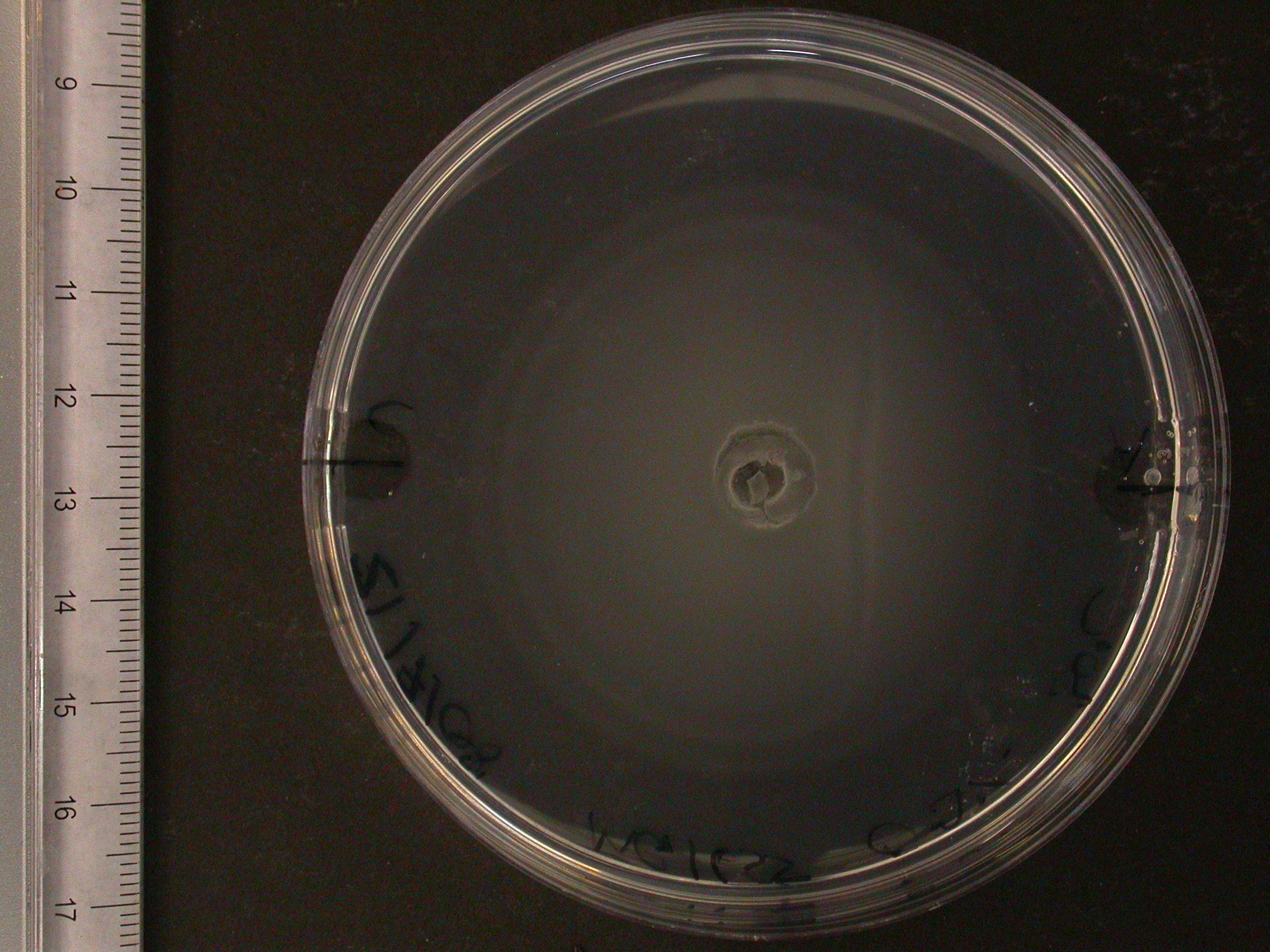
Swimming agar assay
The experiment from 21 July 2008 was repeated. The aspartate solution was added eigth hours before the inoculation. The Results for showed that both strains were actually repelled by aspartate, possibly because the concentration was too high. Aspartate is the well on the right, and the control (water) is on the left.
| 
| 
| 
|
|---|
| MC1000 25oC | MC1000 30oC | MG1655 25oC | MG1655 30oC |
|---|
BioBrick Transformation
The cells transformed with BioBrick were grown on ampicillin plates as if transformed are able to do so, however there was no growth of bacteria except when pUC19 was used. This highlights some problems with the DNA and not the actual transformation. The experiment was repeated with larger amounts of TE buffer around spot and with a different BioBrick. Results see weekly summary.
Bead experiment 1
The bead we had focused on overnight (see 22 July 2008) had gone!! There were observed lots of beads clumping together, areas of dead/non-motile bacteria and also filamentous bacteria (possible conaminant). There were also areas of still motile bacteria. The bacteria had travelled 40 % of way across bridge, thus a smaller bridge was suggested.
 "
"