Team:Chiba/Project
From 2008.igem.org
| Home | The Team | The Project | Parts Submitted to the Registry | Reference | Notebook | Acknowledgements |
|---|
Abstract
E.coli time manager
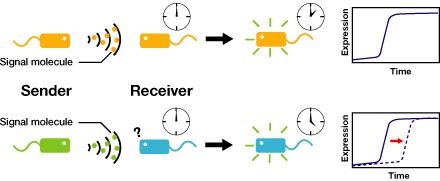
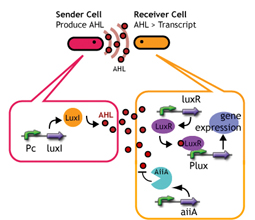
We are constructing delay switches to control/preset the timing of target gene expression. Our project uses two classes of bacteria: senders and receivers. Senders produce signaling molecules, and receivers are activated only after a particular concentration of this molecule is reached. The combinatorial use of senders/receivers allows us to make a‘switching consortium’which activates different genes at the preset times.
As signaling molecules, we utilize molecules associated with Quorum sensing, a phenomenon that allows bacteria to communicate with each other. Although different quorum sensing species have slightly different signaling molecules, these molecules are not completely specific to their hosts and cross-species reactivity is observed (1),(2). Communication using non-endogenous molecules is less sensitive than the original, and requires a higher signal concentration to take effect. This reduced sensitivity results in the slower activation of receivers, thus creating a system in which different receivers are activated after different amounts of time following signaling molecule release.
Introduction
Many electronic devices we use in our daily lives have the ability to keep track of time. For example, a VCR is able to record a TV program at a pre-set time, and a microwave automatically stops heating after a set amount of time. automatically stop heating when the right time comes. Using these temporal pre-programming functions, we have been liberated from either staying up late to watch a European soccer game or from worrying about our popcorn being burned black while yelling and shouting to the match we have videotaped. In this way, the timer function has revolutionized our lifestyle.
We thought the same applies to the biotechnology; we would like to freely implement the 'timer switches” to various biological functions, preferably both independently and in parallel format. These “functions” include sensors, synthesizers, or degraders of bioactive compounds/ materials, transportation and secretion machineries, communications, getting/ sticking together, proliferation and cell death. If successful, we will be able to program exceedingly more complex complex behaviors in cellular systems.
As one of the thousands of possible applications, we are trying to construct a temporal imaging system using E. coli 'ink' that differ not in color but the 'timing' at which they are a certain color (fig). Over time, parts of images (or characters) are getting visible one by one, making animated message/ picture. If the coloration process proceeds to completion, the message is obscured. Only when the message is observed at the correct timing during the coloration process is any useful information obtained. Such a system should be useful for communication security: we can convey our message to only those that know the exact moment they should take a look. After a while, the message is gone and cannot be retrieved.
Project Design
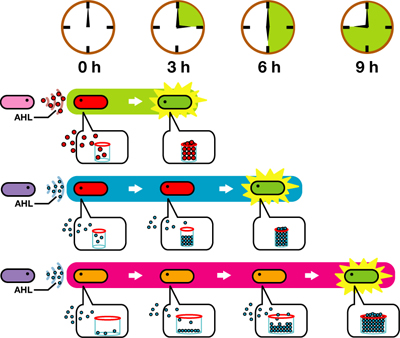
We designed a "switching consortium" that works like a water clock (Water clock-wikipedia.en). Here is how it works (Fig X).
1. Sender cells slowly generate signal molecules AT A CONSTANT RATE. The signal molecules are non-degradable so they get accumulated (linearly) over time.
2. Receivers detect the signal molecules and then activate the genetic switch to the on state, but only when the signal concentration reaches the switching threshold of a receiver. Receivers are activated by different signaling molecules at different rates. In this way, the entire system behaves like a delay switch sequence.
3. Either by changing the receiver sensitivity or rate of signal accumulation, one can freely control the delay length of the individual switches. Using switches with various times-of-delay, one can sequentially activate many different cellular functions.
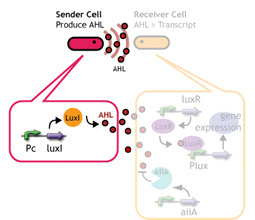
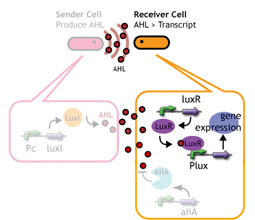
Signaling System
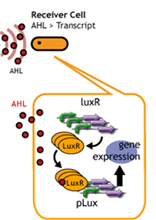
In this project, we use acylated homoserine lactones (AHLs), signaling molecules used for quorum sensing in gram negative bacteria. Senders express LuxI or similar enzymes, which catalyze the production of AHLs, under the control of a constitutive (Tet) promoter. Each cell thus generates AHL more or less at a constant rate. AHL can freely permeate cell membranes and are detected by neighboring cells. Receivers constitutively express LuxR proteins (or a similar ortholog), the protein that detects AHL concentrations. When AHLs bind LuxR proteins, the AHL-LuxR complex activates the Lux promoter. The threshold [AHL] at which switching occurs is determined by the affinity of AHL for the particulr LuxR ortholog.(3),(4). (more about quorum sensing)
System Design
|
プランA: Cross-talk
|
プランB: Tuning
|
List of Experiments
plan A Crosstalk
|
|
|
more about Quorum Sensing Cross-talk
plan B Tuning
|
|
Copy number of Receiver Plasmid
English:
日本語:レシーバーのコピーナンバーを減らすことで、GFPが確認できるまでの時間を延長する。
コピーナンバーが少なくなれば、LuxRの合成量は減少する。
LuxRの合成量が減少すれば、AHLを受け取るLuxRは従来より減ってしまう。
そのため、プロモーターの活性化は遅くなり、GFPの発現量は減る。
したがって、コピーナンバーが多いレシーバーより、GFPが確認できるまでの時間を延長させることができる。
(杉山)
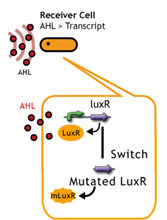
LuxR/Plux mutants show
English:
日本語:私たちはmutated Receiverを用いることで、AHL感受性の違う2種類(WTと変異体)のレシーバーを用意し、delay-timeのバリエーションを増やした。
- a greater response to 3OC6HSL[http://authors.library.caltech.edu/5553/ (3)]
- a increase in sensitivity to 3OC12HSL[http://mic.sgmjournals.org/cgi/content/abstract/151/11/3589 (4)].
Receiver experiments details
-->
LuxR mutant (Under construction)
レシーバータンパク質であるLuxRに変異を入れることで応答感度を上下させる(5),(6)
Demo Experiments
Demo ~Senders~
一番時差が見られたSender遺伝子のLuxIとLasIをつかってデモ実験を行った。
LuxIおよびLasIの遺伝子がそれぞれ組み込まれた大腸菌(XL10G)と、
LuxRの遺伝子が組み込まれた大腸菌(BW)を液体培養したものを 液量1:1で混ぜて、それらのGFPが発現するのを目視で観測した。
Results
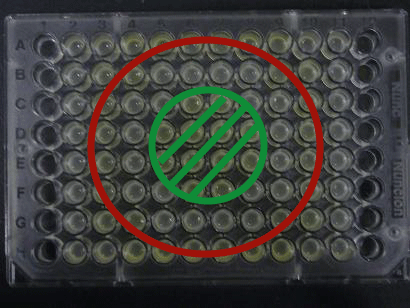
Green region: sender=LuxI (100 uL), Red circular region: sender=Las I (100uL).
Receiver=LuxR (???? uL)
--toyota
-->more about Demo experiments detail
Demo ~Receivers~
English:
日本語:
固体培地中にセンダー[http://partsregistry.org/Part:BBa_S03623 BBa_S03623],(Ptet-LuxI) を混ぜ、固体培地表面にレシーバーのコロニーをN.Cフィルターで移す。
センダーの作るAHLは培地中を移動し、表面のレシーバーがAHLを一定濃度感知すればGFPを発現
する。一種のセンダーに対し、様々な種類のレシーバーを用いることで時間差が生じることを確認する。
用いるレシーバーは・・・
・シグナル自体を分解するAiia を利用する
・レシーバーの遺伝子回路を含むプラスミドのコピーナンバーの変化
・レシーバータンパク質であるLuxRに変異を入れる
・確認の仕方
N.Cフィルターをはった個体培地を37°Cで培養し、時間(30min?)ごとにUVをあててGFPが見えるかチェックする。
香取
Results
--->more about Demo experiments detail
Conclusion
シグナル分子を3OC6HSLから3OC12HSLに変更することで、LuxRの応答時間を二時間遅らせることができた.
以下の実験を行うことで、Delayed Timeのバリエーションがもっと増えるだろう。
- Sender実験とReceiver実験で時間差を作り出すことが出来たものを組み合わせる。
- LuxRのミューテーション。
- ポジティブフィードバックループを利用する。
--Yoshimi 11:09, 29 October 2008 (UTC)
references
- [http://www3.interscience.wiley.com/journal/119124142/abstract M.K Winson et al.:Construction and analysis of luxCDABE-based plasmid sensors for investigating N-acyl homoserine lactone-mediated quorum sensing.FEMS Microbiology Letters 163 (1998) 185-192]
- [http://partsregistry.org/Part:BBa_F2620:Specificity BBa_F2620:Specificity]
- [http://www.pnas.org/content/100/suppl.2/14549.full Michiko E. Taga. Bonnie L.Bassler.:Chemical communication among bacteria.PNAS.November 25, 2003,100.suppl.2]
- [http://arjournals.annualreviews.org/doi/abs/10.1146/annurev.micro.55.1.165?url_ver=Z39.88-2003&rfr_id=ori:rid:crossref.org&rfr_dat=cr_pub%3dncbi.nlm.nih.gov Melissa B. Miller and Bonnie L. Bassler.:QUORUM SENSING IN BACTERIA.annurev.micro.55.1.165.2001]
- [http://authors.library.caltech.edu/5553/ C. H. Collins.et al.:Directed evolution of Vibrio fischeri LuxR for increased sensitivity to a broad spectrum of acyl-homoserine lactones.Mol.Microbiol.2005.55(3).712–723]
- [http://mic.sgmjournals.org/cgi/content/abstract/151/11/3589 B. Koch. et al.:The LuxR receptor: the sites of interaction with quorum-sensing signals and inhibitors.Microbiology 151 (2005),3589-3602]
| Home | The Team | The Project | Parts Submitted to the Registry | Reference | Notebook | Acknowledgements |
|---|
 "
"