Team:ESBS-Strasbourg/Parts
From 2008.igem.org
(Difference between revisions)
(→Toolbox for modular transcription factors) |
|||
| Line 8: | Line 8: | ||
==Toolbox for modular transcription factors== | ==Toolbox for modular transcription factors== | ||
| + | [[image:ESBS_TF.jpg|thumb|right]] | ||
===DNA binding domains=== | ===DNA binding domains=== | ||
* '''tetR''' [http://partsregistry.org/wiki/index.php?title=Part:BBa_K105003 BBa_K105003] | * '''tetR''' [http://partsregistry.org/wiki/index.php?title=Part:BBa_K105003 BBa_K105003] | ||
| Line 31: | Line 32: | ||
<br> | <br> | ||
<br> | <br> | ||
| + | |||
==Toolbox for modular promoters== | ==Toolbox for modular promoters== | ||
===Minimal promoters of different strength=== | ===Minimal promoters of different strength=== | ||
Revision as of 23:00, 29 October 2008
Parts submitted
Following you can find a summary of our contribution to the registry.
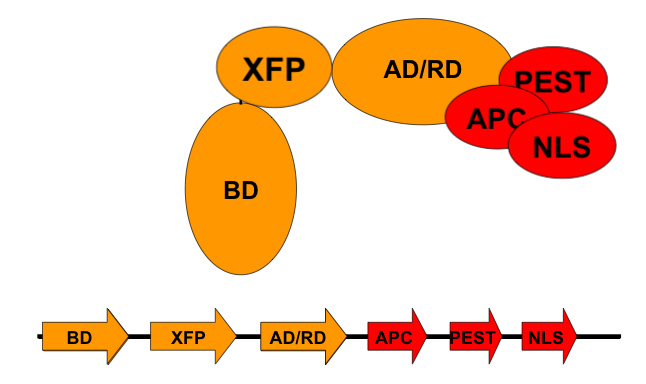
Toolbox for modular transcription factors
DNA binding domains
- tetR [http://partsregistry.org/wiki/index.php?title=Part:BBa_K105003 BBa_K105003]
- cI [http://partsregistry.org/wiki/index.php?title=Part:BBa_K105004 BBa_K105004]
- lexA [http://partsregistry.org/wiki/index.php?title=Part:BBa_K105005 BBa_K105005]
- lacI [http://partsregistry.org/wiki/index.php?title=Part:BBa_K105006 BBa_K105006]
- Gal4 [http://partsregistry.org/wiki/index.php?title=Part:BBa_K105007 BBa_K105007]
Activation domain
- VP16 [http://partsregistry.org/wiki/index.php?title=Part:BBa_K105001 BBa_K105001]
Repression domain
- TUP1 [http://partsregistry.org/wiki/index.php?title=Part:BBa_K105002 BBa_K105002]
Fluorescent markers
- CFP [http://partsregistry.org/wiki/index.php?title=Part:BBa_K105009 BBa_K105009]
- mCherry [http://partsregistry.org/wiki/index.php?title=Part:BBa_K105010 BBa_K105010]
- mOrange [http://partsregistry.org/wiki/index.php?title=Part:BBa_K105011 BBa_K105011]
Junctions
- linker [http://partsregistry.org/wiki/index.php?title=Part:BBa_K105012 BBa_K105012]
Tags
- cin8 - cell cycle dependent degradation [http://partsregistry.org/wiki/index.php?title=Part:BBa_K105013 BBa_K105013]
- hsl1 – cell cycle dependent degradation [http://partsregistry.org/wiki/index.php?title=Part:BBa_K105015 BBa_K105015]
=> all these parts are in our new plasmid derivate [http://partsregistry.org/wiki/index.php?title=Part:BBa_K105000 pSB1A2_fusion]
Toolbox for modular promoters
Minimal promoters of different strength
- cyc100 [http://partsregistry.org/wiki/index.php?title=Part:BBa_K105027 BBa_K105027]
- cyc70 [http://partsregistry.org/wiki/index.php?title=Part:BBa_K105028 BBa_K105028]
- cyc43 [http://partsregistry.org/wiki/index.php?title=Part:BBa_K105029 BBa_K105029]
- cyc28 [http://partsregistry.org/wiki/index.php?title=Part:BBa_K105030 BBa_K105030]
- cyc16 [http://partsregistry.org/wiki/index.php?title=Part:BBa_K105031 BBa_K105031]
Operator sites
- tetR operator [http://partsregistry.org/wiki/index.php?title=Part:BBa_K105020 BBa_K105020]
- cI operator [http://partsregistry.org/wiki/index.php?title=Part:BBa_K105021 BBa_K105021]
- lexA operator [http://partsregistry.org/wiki/index.php?title=Part:BBa_K105022 BBa_K105022]
- lacI operator [http://partsregistry.org/wiki/index.php?title=Part:BBa_K105023 BBa_K105023]
- Gal4 operator [http://partsregistry.org/wiki/index.php?title=Part:BBa_K105024 BBa_K105024]
Yeast selection markers
- Leu2 - autotrophy marker [http://partsregistry.org/wiki/index.php?title=Part:BBa_K105016 BBa_K105016]
- Ura3 - autotrophy marker [http://partsregistry.org/wiki/index.php?title=Part:BBa_K105017 BBa_K105017]
Remakes of good, but not available BioBricks
- Gal1 promoter [http://partsregistry.org/wiki/index.php?title=Part:BBa_K105026 BBa_K105026]
- Adh1 terminator [http://partsregistry.org/wiki/index.php?title=Part:BBa_K105025 BBa_K105025]
 "
"