Team:ESBS-Strasbourg/Results
From 2008.igem.org
(→Fusion vector) |
(→Assembly) |
||
| Line 22: | Line 22: | ||
==Assembly== | ==Assembly== | ||
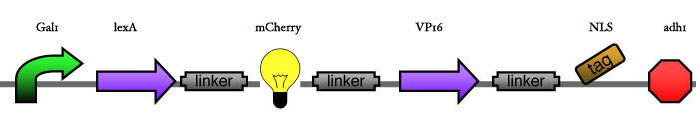
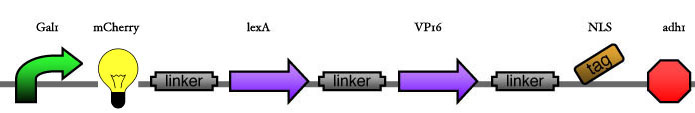
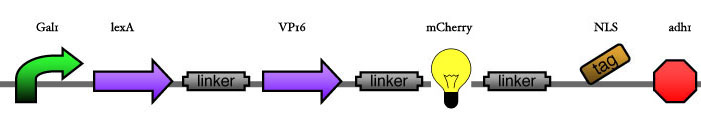
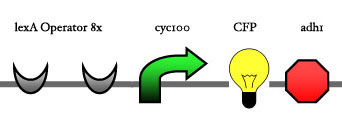
| - | Our assembly strategy, described in the measurement section, was followed in order to figure out the optimal order of single parts. Following are monitored the constructs we have assembled. We refer to the measurement section if their usefulness is unclear. Their function has merely to be demonstrated through transformation into yeast. Various other meta-assemblies (not shown) of the measurement have been constructed. | + | Our assembly strategy, described in the [https://2008.igem.org/Team:ESBS-Strasbourg/measurements measurement] section, was followed in order to figure out the optimal order of single parts. Following are monitored the constructs we have assembled. We refer to the measurement section if their usefulness is unclear. Their function has merely to be demonstrated through transformation into yeast. Various other meta-assemblies (not shown) of the measurement have been constructed. |
<br> | <br> | ||
[[Image:ESBS-tester11.JPG]]<br> | [[Image:ESBS-tester11.JPG]]<br> | ||
| Line 33: | Line 33: | ||
[[Image:ESBS-tester44.JPG]]<br> | [[Image:ESBS-tester44.JPG]]<br> | ||
<br> | <br> | ||
| + | |||
==Theory & Modeling== | ==Theory & Modeling== | ||
We have developed theoretically a project of a cell cycle dependent toggle switch. Thereby we have introduced new ideas to the synthetic biology community through a radical usage of the BioBrick concept. In first instance, we have started to divide coding sequences as far as possible into their functional domains. This allows us to build up easily components as transcription factors and repressors with wished properties. Secondly, we have also divided all regulatory sequences. Operator, promoter and Kozak sequence are thus available separately, allowing assembling regulatory elements with desired activation and transcription rates. We have also adapted the BioBrick concept for auxotrophic marker sequences, which are of rather technical nature. This radical usage could, on the one hand allow to build up more precise systems. On the other hand it could facilitate the construction of systems since an automated BioBrick assembly can be used to even higher degree.<br> | We have developed theoretically a project of a cell cycle dependent toggle switch. Thereby we have introduced new ideas to the synthetic biology community through a radical usage of the BioBrick concept. In first instance, we have started to divide coding sequences as far as possible into their functional domains. This allows us to build up easily components as transcription factors and repressors with wished properties. Secondly, we have also divided all regulatory sequences. Operator, promoter and Kozak sequence are thus available separately, allowing assembling regulatory elements with desired activation and transcription rates. We have also adapted the BioBrick concept for auxotrophic marker sequences, which are of rather technical nature. This radical usage could, on the one hand allow to build up more precise systems. On the other hand it could facilitate the construction of systems since an automated BioBrick assembly can be used to even higher degree.<br> | ||
Revision as of 01:22, 30 October 2008
Outcome
Our goal was to construct a cell cycle dependent toggle switch in eukaryotic cells. Unfortunately, we could reach this goal not completely because we did not have enough time to assemble the system at the bench. Following are described the meta-results we produced as well as our contribution to the iGEM community.
Fusion vector
We have established a new vector, which allows making fusion proteins of two standard BioBrick parts. This vector is characterized by the absence of two base pairs. In all other details it is identical to the vector [http://partsregistry.org/Part:pSB1A3 pSB1A2]. The base pairs, which have been removed, are the one downstream of the Xba1 cleaving site and the one upstream of the Spe1 site. Consequently, an assembly of two parts will have 6 base pairs (ACTAGA) in between the part’s sequences, instead of 8 base pairs. Thus, a frame shift will be avoided. The consequent amino acids between the two parts are threonine and arginine. The construction of this vector is based on the technical note of Ira Phillips and Pamela Silver (Phillips, IE; Silver, PA; 2006). The propositions of the document should be followed if parts want to be cloned into this vector. All our parts submitted, described following were cloned into this vector.
BioBricks
Our team submitted 27 new parts to the Registry, which will serve further generations of iGEM teams as well as the whole synthetic biology community in the future. Since our system was envisaged to work in yeast among these parts are numerous eukaryotic and yeast specific parts which will be of high value for the underrepresented yeast parts section of the Registry. 16 parts are completely new sequences, which have never been registered in this or related form. The most important of these BioBricks are described shortly following:
- The activation domain VP16 as well as the repression domain Tup1 allow regulation of the eukaryotic transcription machinery
- A linker sequence allows constructing fusion proteins between different domains, which should keep their individual mobility to a certain degree
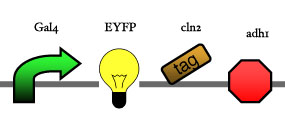
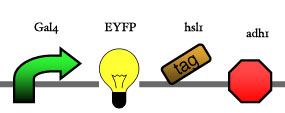
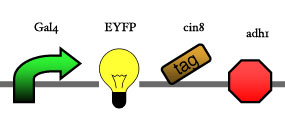
- Cin8 and hsl1 are cell cycle specific degradation tags, which can be used as income for specific time frames of the cell cycle
- Leu2 and Ura3 are auxotrophic markers for the selection of successfully transformed yeast
- Cyc1 Promoter of various strength as well as specific operators allow to construct individual regulatory sequences
The remaining 11 parts correspond to sequences, which have been already registered in a similar form. In order to use them for our project we have liberated them from start and stop codons. Thus, an unlimited use of these BioBricks to construct fusion proteins is now possible. Among them, are the most important fluorescence proteins and prokaryotic binding domains.
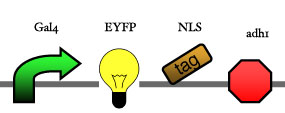
Assembly
Our assembly strategy, described in the measurement section, was followed in order to figure out the optimal order of single parts. Following are monitored the constructs we have assembled. We refer to the measurement section if their usefulness is unclear. Their function has merely to be demonstrated through transformation into yeast. Various other meta-assemblies (not shown) of the measurement have been constructed.
Theory & Modeling
We have developed theoretically a project of a cell cycle dependent toggle switch. Thereby we have introduced new ideas to the synthetic biology community through a radical usage of the BioBrick concept. In first instance, we have started to divide coding sequences as far as possible into their functional domains. This allows us to build up easily components as transcription factors and repressors with wished properties. Secondly, we have also divided all regulatory sequences. Operator, promoter and Kozak sequence are thus available separately, allowing assembling regulatory elements with desired activation and transcription rates. We have also adapted the BioBrick concept for auxotrophic marker sequences, which are of rather technical nature. This radical usage could, on the one hand allow to build up more precise systems. On the other hand it could facilitate the construction of systems since an automated BioBrick assembly can be used to even higher degree.
Our system has proven to be work in silico with a wide variety of parameters. Therefore, we are optimistic in regard to its practical functioning. The information given on this website should allow future generation to continue this work.
 "
"