Team:Hawaii/Notebook/2008-08-31
From 2008.igem.org
(Difference between revisions)
(→Construction of p+r (cont.) and re-replacement of BB-pRL1833a MCS) |
(margaret) |
||
| Line 14: | Line 14: | ||
:* Transformed into DH5α cells | :* Transformed into DH5α cells | ||
::* Plated BB-pRL1383a+J33207 on selective media with and without IPTG to verify if IPTG is necessary for blue/white screening in this strain | ::* Plated BB-pRL1383a+J33207 on selective media with and without IPTG to verify if IPTG is necessary for blue/white screening in this strain | ||
| + | |||
| + | ===Construction of Broad-Host-Range Plasmid Parts=== | ||
| + | <strong>Margaret</strong> | ||
| + | |||
| + | [[Image:re_digest_8_31_08.jpg|right|thumb|150px|Restriction digest of rep and P1 lytic regions.]] | ||
| + | :*gel of yesterday's re-digest, lane 3 was cleaned using gel purification spin columns and stored in -20°C. Lanes 7-10 were extracted and cleaned as well. | ||
| + | :*Ligation: using T4 ligase, 10X buffer (from the same lot as the enzyme, it was aliquoted today). 2 hour incubation at room temperature. The reaction was placed in 4°C in case transformation does not yield colonies. | ||
| + | |||
| + | {|class=wikitable border=1 align=center | ||
| + | !insert | ||
| + | !vector | ||
| + | |- | ||
| + | |1 pSB1A3 cut with X,P(de-P0<sub>4</sub><sup>2-</sup>)18ng | ||
| + | |3 rep (PCR product) X,P 82ng | ||
| + | |- | ||
| + | |1pSB1A3 cut with X,P(de-P0<sub>4</sub><sup>2-</sup>)35.6ng | ||
| + | |3P1 lytic region X,P 64.4ng | ||
| + | |- | ||
| + | |3pSB1A3 cut with X,P(de-P0<sub>4</sub><sup>2-</sup>)66.2ng | ||
| + | |1 rep (PCR product) X,P 33.8ng\ | ||
| + | |} | ||
| + | |||
| + | :*Transformation: of ligation products from today into DH5-alpha cells. Everything plated on Amp100 LB plates, pSMC121 used as positive control (plated on SmSp). | ||
| + | |||
| + | |||
| + | |||
= Discussion = | = Discussion = | ||
Revision as of 03:24, 1 September 2008
| Projects | Events | Resources | ||
|---|---|---|---|---|
| Sponsors | Experiments | Milestones | Protocols | |
| Notebook (t) | Meetings (t) |
Things we did today
Wetlab work
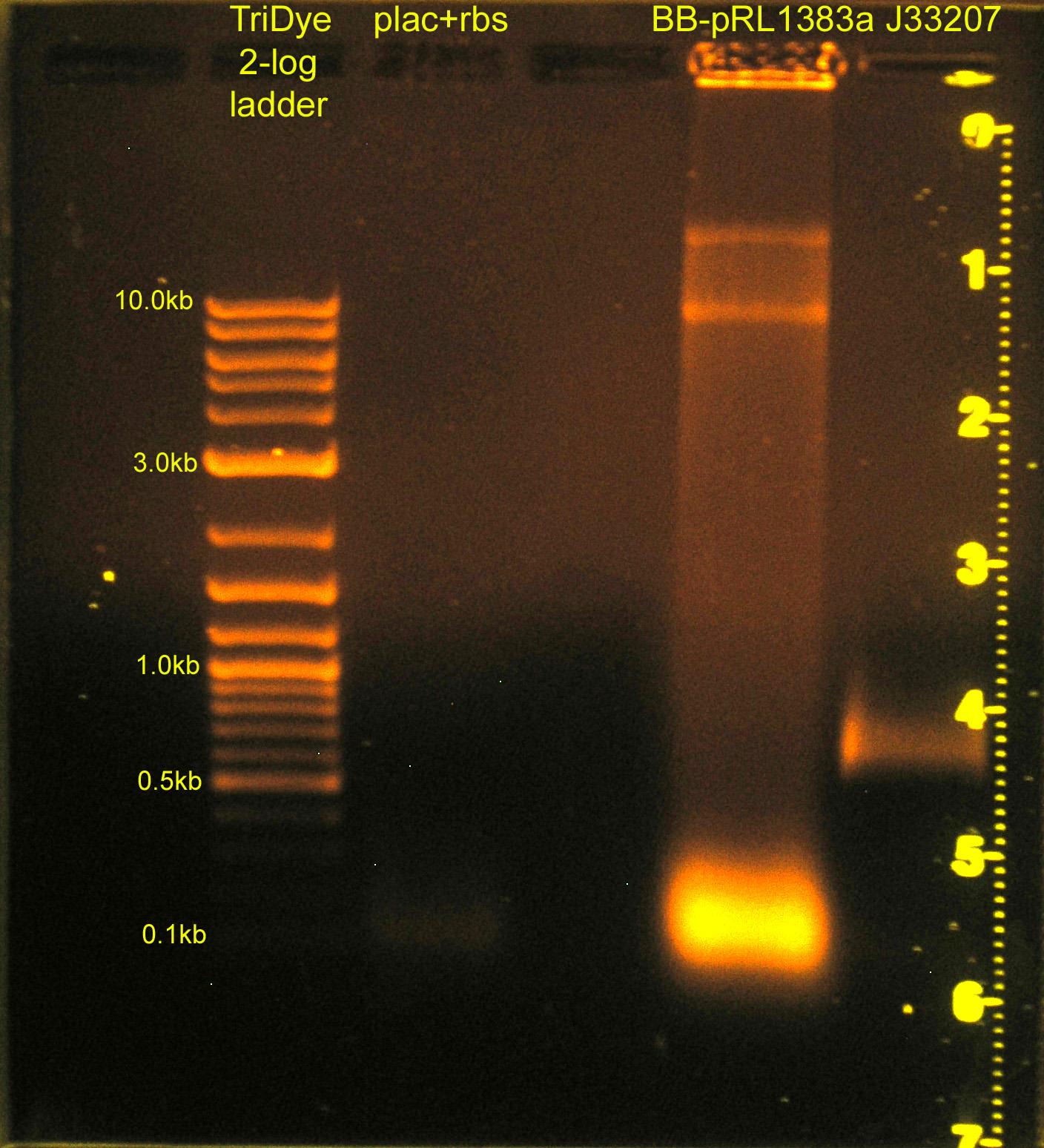
Construction of p+r (cont.) and re-replacement of BB-pRL1833a MCS
- Grace
- Ran RE digests from last night on gel
- plac+rbs band not visible; will ligate using plac and rbs parts (instead of plac+rbs part)
- Extracted bands from gel
- Ligated:
- plac and rbs (B0034) and C0012 vector
- J33207 and BB-pRL1383a
- Transformed into DH5α cells
- Plated BB-pRL1383a+J33207 on selective media with and without IPTG to verify if IPTG is necessary for blue/white screening in this strain
Construction of Broad-Host-Range Plasmid Parts
Margaret
File:Re digest 8 31 08.jpg
Restriction digest of rep and P1 lytic regions.
- gel of yesterday's re-digest, lane 3 was cleaned using gel purification spin columns and stored in -20°C. Lanes 7-10 were extracted and cleaned as well.
- Ligation: using T4 ligase, 10X buffer (from the same lot as the enzyme, it was aliquoted today). 2 hour incubation at room temperature. The reaction was placed in 4°C in case transformation does not yield colonies.
| insert | vector |
|---|---|
| 1 pSB1A3 cut with X,P(de-P042-)18ng | 3 rep (PCR product) X,P 82ng |
| 1pSB1A3 cut with X,P(de-P042-)35.6ng | 3P1 lytic region X,P 64.4ng |
| 3pSB1A3 cut with X,P(de-P042-)66.2ng | 1 rep (PCR product) X,P 33.8ng\ |
- Transformation: of ligation products from today into DH5-alpha cells. Everything plated on Amp100 LB plates, pSMC121 used as positive control (plated on SmSp).
Discussion
Quote of the Day
History is the only laboratory we have in which to test the consequences of thought. - Étienne Gilson
[http://manoa.hawaii.edu/  ][http://manoa.hawaii.edu/ovcrge/
][http://manoa.hawaii.edu/ovcrge/  ][http://www.ctahr.hawaii.edu
][http://www.ctahr.hawaii.edu  ]
]
 "
"