Team:KULeuven/13 September 2008
From 2008.igem.org
(wet lab) |
|||
| Line 1: | Line 1: | ||
| + | {{:Team:KULeuven/Tools/Styling}} | ||
{{:Team:KULeuven/Tools/Header}} | {{:Team:KULeuven/Tools/Header}} | ||
| + | |||
| + | {{:Team:KULeuven/Tools/New_Day/Date_Retriever}} | ||
== Lab Work == | == Lab Work == | ||
| Line 25: | Line 28: | ||
== Remarks == | == Remarks == | ||
| - | |||
| - | |||
| - | |||
Revision as of 19:43, 28 October 2008
| << return to notebook | return to homepage >> | ||
| < previous friday | ← yesterday | tomorrow → | next monday > |
Contents |
Lab Work
Wet Lab
We miniprepped two cultures today: K145272 and K145013+B0015.
We also did a new PCR test on the electroporations of the day before yesterday (but different colonies). We found that colony 3 of B0014+B0033+K145151+B0015 is probably correct. The other ones were still wrong though.
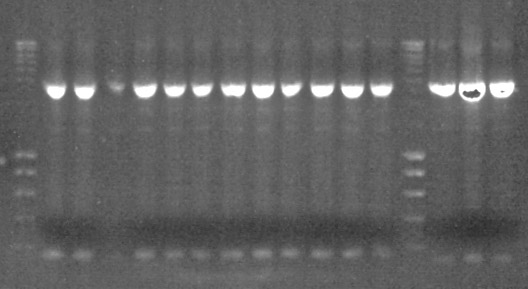
We did step 2 of the PCR to construct T7 DNA polymerase with UmuD tag - but without purifying the mix obtained in step 1. And check it out, it seemed to have worked! Hurrah! Take a look at the picture on the right and you will see that we had a band at the correct height (2750 bp). Thank you professor Robben.
Apart from that, we also electroporated J23116+B0034 and K145275; and we made a ligation mix of C0062+B0015 and I712074+J23078.
We set up some digests. After 2 hours we put them on gel to see if they were properly cut. Unfortenately they weren't. We let them stand overnight and hope that they will be digested by tomorrow (it were the last bits of enzym, so maybe they don't work anymore):
 "
"