Team:KULeuven/Model/Filter
From 2008.igem.org
Contents |
Filter
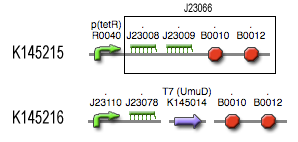
Position in the system
The filter is positioned immediately after the input, because its job is to filter out possible noise signals or background signals that aren't caused by the "desease". It is the starting piece of the whole system, situated before the invertimer- and the reset-subsystem.
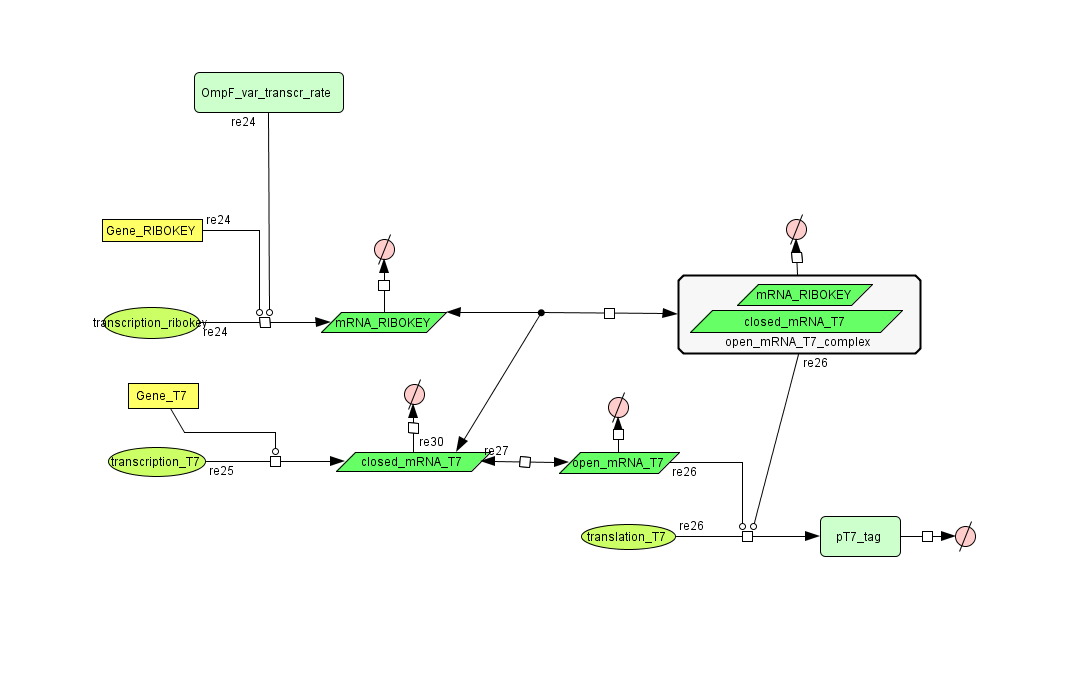
Describing the system
see also: Project:Filter
ODE's
Parameters
| Name | Value | Comments | Reference |
|---|---|---|---|
| Degradation rates | |||
| dpT7 tag | 0.00155 s-1 | UmuD tag added to speed up degradation of otherwise too stable T7 polymerase | [http://www.openwetware.org/wiki/IGEM:Tsinghua/2007/Projects/RAP#Model_and_simulation link] |
| dmRNA RIBOKEY | 0.00462 s-1 | estimate: because this RNA isn't translated, it degrades faster | [http://www.pubmedcentral.nih.gov/picrender.fcgi?artid=124983&blobtype=pdf link] |
| dclosed mRNA T7 | 0.00462 s-1 | estimate: because this mRNA isn't translated, it degrades faster | [http://www.pubmedcentral.nih.gov/picrender.fcgi?artid=124983&blobtype=pdf link] |
| dopen mRNA T7 | 0.00231 s-1 | [http://www.pubmedcentral.nih.gov/picrender.fcgi?artid=124983&blobtype=pdf link] | |
| dopen mRNA T7 complex | 0.00231 s-1 | [http://www.pubmedcentral.nih.gov/picrender.fcgi?artid=124983&blobtype=pdf link] | |
| Equilibrium constants | |||
| Keq 1 | 0,015 [M] | between closed and open T7 mRNA, models competition, experimental | [http://parts2.mit.edu/wiki/index.php/Berkeley2006-RiboregulatorsMain link] |
| Keq 2 | 0.0212 [M] | between closed T7 mRNA and key unlocked mRNA complex, models competition, experimental | [http://parts2.mit.edu/wiki/index.php/Berkeley2006-RiboregulatorsMain link] |
| Rate constants | |||
| kdis | 0.00416 s-1 | estimate derived from experimental values | [http://parts2.mit.edu/wiki/index.php/Berkeley2006-RiboregulatorsMain link] |
| kcomplex | 0.00237 s-1 | estimate derived from experimental values | [http://parts2.mit.edu/wiki/index.php/Berkeley2006-RiboregulatorsMain link] |
| kclosed | 500 s-1 | estimate derived from experimental values | [http://parts2.mit.edu/wiki/index.php/Berkeley2006-RiboregulatorsMain link] |
| kopen | 7.5 s-1 | estimate derived from experimental values | [http://parts2.mit.edu/wiki/index.php/Berkeley2006-RiboregulatorsMain link] |
| ktranslation | 0.167 s-1 | lock defined translation rate for T7 RNA pol | [http://partsregistry.org/Part:BBa_B0032 link] |
| Transcription rates | |||
| TetR_var_transcr_rate | p(TetR) dependent | (RiboKey) between 5E-5 and 0.0125 s-1 ~ [aTc] | |
| kmRNA T7 | 0,0011 s-1 | weak constitutive promoter J23109 | [http://partsregistry.org/Part:BBa_J23109 link] |
Remark: The key-lock system has been enhanced to 0.3%-14% (new parameters have been added)
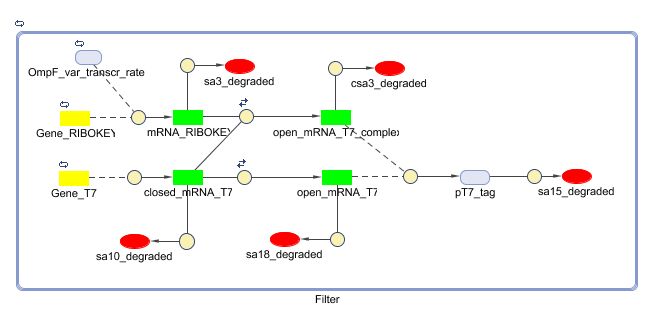
Models
CellDesigner (SBML file)
Matlab (SBML file)
Simulations
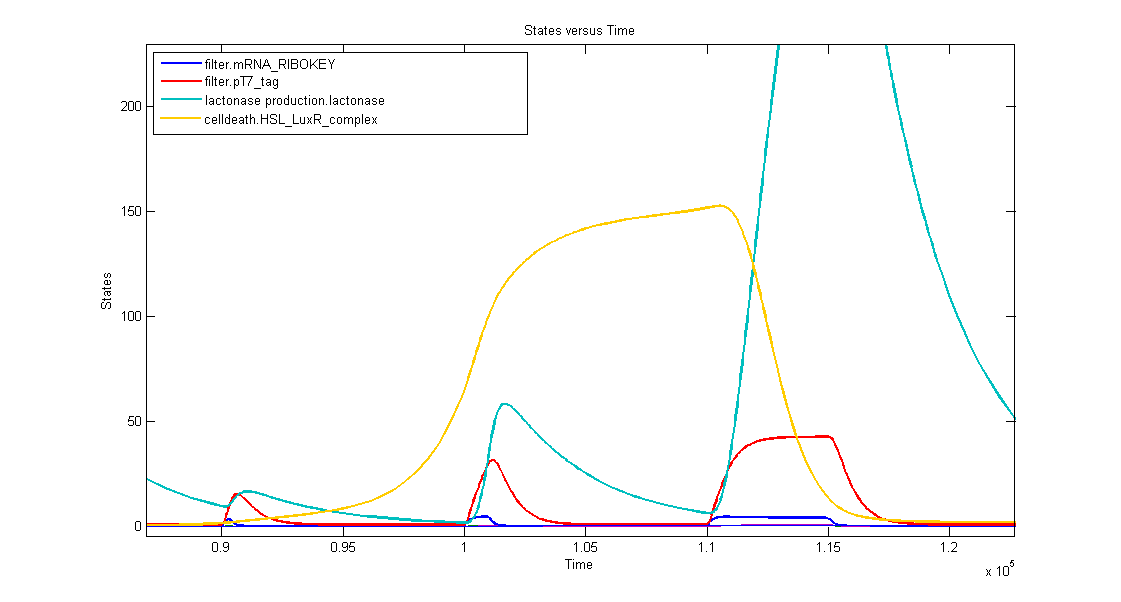
1. AND gate of the filter
In the simulation we can clearly see this series of events:
- when dark blue(ribokey) starts to increase, red (T7) also starts to increase, giving rise to an increasing amount of lactonase (blue) = AND-GATE.
- when dark blue(ribokey) starts to decrease, red (T7) also starts to decrease, but much slower. The lactonase also starts to decrease, as it should be.
The short lifetime of the ribokey compared to the lifetime of the T7-protein, guarantees that the AND-GATE always works perfectly fine: when there's no more input, the ribokey will rapidly decrease (and disappear) and makes sure that the AND-GATE is not activated anymore, even when the T7-protein is slowly decreasing.
2. Filtering in practice
In the simulation three kinds of inputpulses have been used:
- First pulse: 300s
- small peak of lactonase
- no influence on the timer
- Second pulse: 1000s
- medium peak of lactonase
- influences the timer by levelling the timing capabilities, but it doesn't reset the timer
- Third pulse: 5000s
- huge peak of lactonase
- reset of the timer: amount of complex goes to zero
3. Proof of filtering capacities
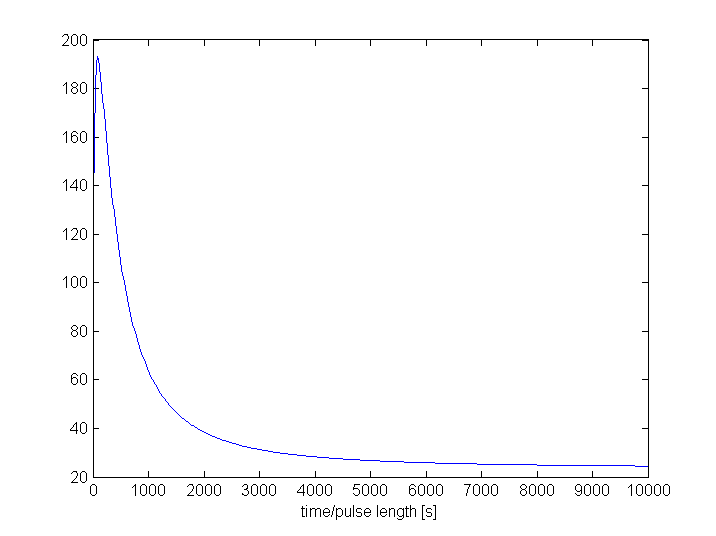
As proof of the filtering capacities, we'll compare the maximum amount of GFP with the maximum amount of Lactonase as a function of the pulse length of TetR. We've chosen Lactonase and GFP because they only differ in the absence or presence of the filter. The used model for the proof consists only of the filter, the output and the lactonase productions subsystems.
The left figures shows us a clear difference between the amount of GFP and lactonase for almost every pulse length. The explanation for the (almost constant) big difference for pulses longer than 1000 seconds is the following: For such long pulses, the filter is fully active such that the only differences between the output and the lactonase production are the limitations by the ribokey and the difference in transcription factors. For a pulse of less than 1000 seconds the filter is not fully active. This results in a filtering effect. If we zoom for only the first 1000 seconds (right figures), we see that the amount of Lactonase remains more or less the same while the amount of GFP increases very fast.


Another way to see the filtering effect is to look at the ratio of GFP with Lactonase. This figure clearly shows the disproportionality between GFP and lactonase for the pulses shorter than 1000 seconds.
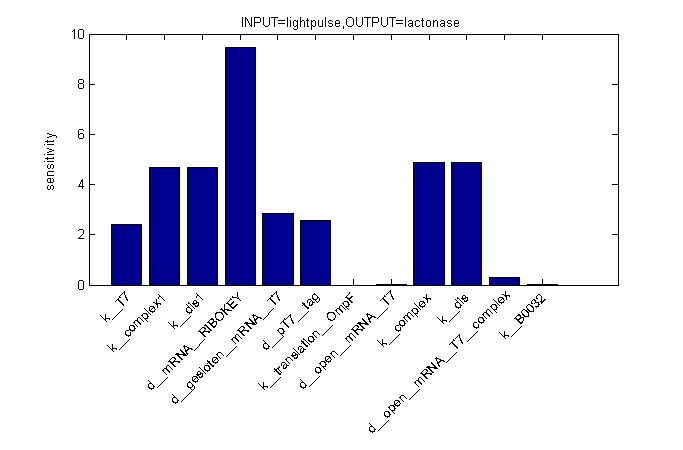
Sensitivity Analysis
References
| [1] | “Berkeley2006-RiboregulatorsMain - IGEM”; http://parts2.mit.edu/wiki/index.php/Berkeley2006-RiboregulatorsMain. |
| [2] | F.J. Isaacs et al., “Engineered riboregulators enable post-transcriptional control of gene expression,” Nat Biotech, vol. 22, Jul. 2004, pp. 841-847. |
| [3] | J.A. Bernstein et al., “Global analysis of mRNA decay and abundance in Escherichia coli at single-gene resolution using two-color fluorescent DNA microarrays,” Proceedings of the National Academy of Sciences of the United States of America, vol. 99, Jul. 2002, pp. 9697–9702. |
| [4] | “IGEM:Tsinghua/2007/Projects/RAP - OpenWetWare”; http://www.openwetware.org/wiki/IGEM:Tsinghua/2007/Projects/RAP#Model_and_simulation. |
| [5] | M. Gonzalez et al., “Lon-mediated proteolysis of the Escherichia coli UmuD mutagenesis protein: in vitro degradation and identification of residues required for proteolysis,” Genes Dev., vol. 12, Dec. 1998, pp. 3889-3899. |
| [6] | M. Bon, S.J. McGowan, and P.R. Cook, “Many expressed genes in bacteria and yeast are transcribed only once per cell cycle,” FASEB J., vol. 20, Aug. 2006, pp. 1721-1723. |
| [7] | “Part:BBa B0032 - partsregistry.org”; http://partsregistry.org/Part:BBa_B0032. |
| [8] | “Part:BBa J23109 - partsregistry.org”; http://partsregistry.org/Part:BBa_J23109. |
| [9] | J.P. McDonald et al., “Regulation of UmuD cleavage: role of the amino-terminal tail,” Journal of Molecular Biology, vol. 282, Oct. 1998, pp. 721-730. |
 "
"