Team:NYMU-Taipei/Project/pH Sensor
From 2008.igem.org
(Difference between revisions)
Blackrabbit (Talk | contribs) |
(→NhaA promoter under 0uM, 150uM, 300uM sodium and pH 3.0, 7.0, 8.5) |
||
| (2 intermediate revisions not shown) | |||
| Line 49: | Line 49: | ||
* [http://www.sciencedirect.com/science?_ob=ArticleURL&_udi=B6T1S-4CVR4C8-1&_user=10&_rdoc=1&_fmt=&_orig=search&_sort=d&view=c&_acct=C000050221&_version=1&_urlVersion=0&_userid=10&md5=5cb90ff8555f4439c804a59f5b397c16 NhaA of Escherichia coli, as a model of a pH-regulated Na+/H+antiporter] (E. Padan etc., 2004) | * [http://www.sciencedirect.com/science?_ob=ArticleURL&_udi=B6T1S-4CVR4C8-1&_user=10&_rdoc=1&_fmt=&_orig=search&_sort=d&view=c&_acct=C000050221&_version=1&_urlVersion=0&_userid=10&md5=5cb90ff8555f4439c804a59f5b397c16 NhaA of Escherichia coli, as a model of a pH-regulated Na+/H+antiporter] (E. Padan etc., 2004) | ||
* [http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pubmed&pubmedid=11133959 Transcription of nhaA, the Main Na+/H+ Antiporter of Escherichia coli, Is Regulated by Na+ and Growth Phase] (Nir Dover and Etana Padan et al.,2001) | * [http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pubmed&pubmedid=11133959 Transcription of nhaA, the Main Na+/H+ Antiporter of Escherichia coli, Is Regulated by Na+ and Growth Phase] (Nir Dover and Etana Padan et al.,2001) | ||
| + | |||
| + | == Experiments == | ||
| + | |||
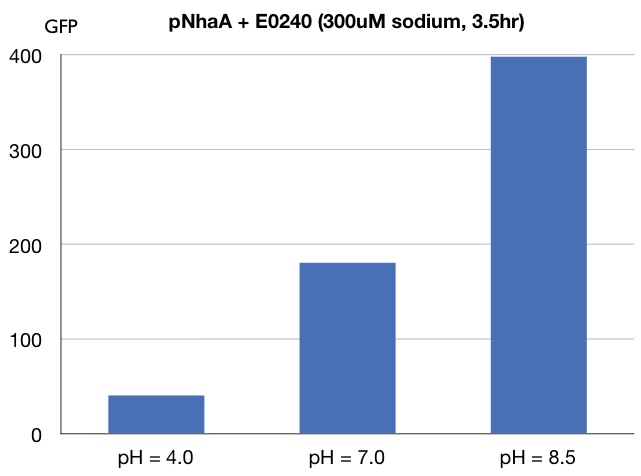
| + | === ''NhaA'' promoter under 300uM sodium and pH 3.0, 7.0, 8.5 === | ||
| + | |||
| + | |||
| + | {| | ||
| + | | | ||
| + | [[Image:PNhaA 20081025 300uM Na 3.5h.png|400px]] | ||
| + | |- | ||
| + | |} | ||
| + | |||
{{:Team:NYMU-Taipei/Footer}} | {{:Team:NYMU-Taipei/Footer}} | ||
Latest revision as of 05:10, 30 October 2008
| Home | Project Overview: | pH Sensor | Attachment | Time Regulation | Waste Removal | Experiments and Parts | About Us |
Contents |
Motivation
- When our system arrives in the intestine (pH is higher), it senses the pH change and starts to work.
- In this subsystem, we are going to create a pH sensor which senses high pH's.
Goal
- The pH sensor can sense a high pH condition and start gene expression.
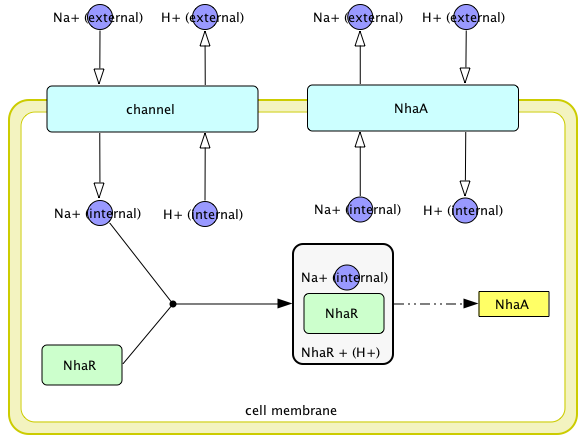
Circuit Design
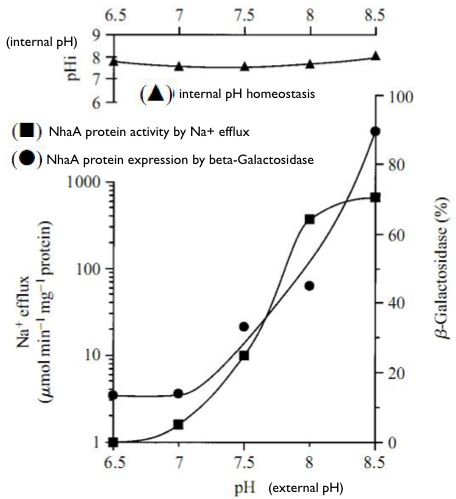
Regulation of nhaA gene expression
- Na1-Induced Transcription of nhaA, Which Encodes an Na1/H1 Antiporter in Escherichia coli, Is Positively Regulated by nhaR and Affected by hns (N. DOVER etc.,1996)
- Molecular physiology of the Na+/H+ antiporter in Escherichia coli (E Padan and S Schuldiner, 1994)
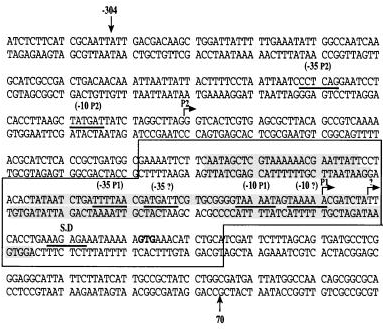
Structure of nhaA gene promoter
- Transcription of nhaA, the Main Na+/H+ Antiporter of Escherichia coli, Is Regulated by Na+ and Growth Phase (Nir Dover and Etana Padan et al.,2001)
- 17,189 -17,488 in E.coli K12 genome from Ecocyc (300 bp upstream of nhaA gene)
00000000011111111112222222222344444444455555555556666666666777777777788888888889 12345678901234567890123456789012345678901234567890123456789012345678901234567890 ACGACAAGCTGGATTATTTTTGAAATATTGGCCTAACAAGCATCGCCGACTGACAACAAATTAATTATTACTTTTCCTAA TTAATCCCTCAGGAATCCTCACCTTAAGCTATGATTATCTAGGCTTAGGGTCACTCGTGAGCGCTTACAGCCGTCAAAAA CGCATCTCACCGCTGATGGCGCAAATTCTTCAATAGCTCGTAAAAAACGAATTATTCCTACACTATAATCTGATTTTAAC GATGATTCGTGCGGGGTAAAATAGTAAAAACGATCTATTCACCTGAAAGAGAAATAAAAA
- G is the start of NhaR binding site
- TTTTAA is the first -35 of P1
- ATGATT is the second -35 of P1
- TAAAAT is the first -10 of P1
- TAAAAA is the second -10 of P1; A in the TAAAAA is the first TSS of P1
- AAGAGA is the S.D. (Shine-Dalgarno) sequence in RBS
- There are 3 nhaA promoter sequences protected by NhaR from DNase I digestion
- AATAGCTCGTAAAAAACGAATTATTCC
- CACTATAATCTGATTTTAACGATG
- CGTGCGGGGTAAAATAGTAAAAACGATCTATTCACCT; T in TTCACCT is the second TSS of P1
- The extracted DNA sequence should include the NhaR binding site
- The NhaR binding site defined in (Nir Dover and Etana Padan et al.,2001) is 120 bp long
- The PCR product derived from primers designed by Henry is 274 bp long
References
- Molecular physiology of the Na+/H+ antiporter in Escherichia coli (E Padan and S Schuldiner, 1994)
- Histidine-226 is part of the pH sensor of NhaA, a Na+/H+ antiporter in Escherichia coli (Y Gerchman etc., 1993)
- Structure of a Na+/H+ antiporter and insights into mechanism of action and regulation by pH (Carola Hunte etc., 2005)
- Na1-Induced Transcription of nhaA, Which Encodes an Na1/H1 Antiporter in Escherichia coli, Is Positively Regulated by nhaR and Affected by hns (N. DOVER etc.,1996)
- Molecular physiology of the Na+/H+ antiporter in Escherichia coli (E Padan and S Schuldiner, 1994)
- NhaA of Escherichia coli, as a model of a pH-regulated Na+/H+antiporter (E. Padan etc., 2004)
- Transcription of nhaA, the Main Na+/H+ Antiporter of Escherichia coli, Is Regulated by Na+ and Growth Phase (Nir Dover and Etana Padan et al.,2001)
 "
"