Team:Paris/August 11
From 2008.igem.org
(→PCR verification/Analysis) |
(→PCR) |
||
| Line 304: | Line 304: | ||
<br> samples : 3µl of PCR products + 2µl of Loading Dye | <br> samples : 3µl of PCR products + 2µl of Loading Dye | ||
<br> migration 30min at 100V, on a '''2%''' agarose gel | <br> migration 30min at 100V, on a '''2%''' agarose gel | ||
| + | |||
| + | |||
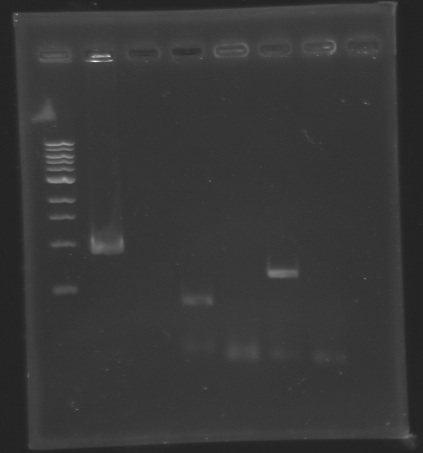
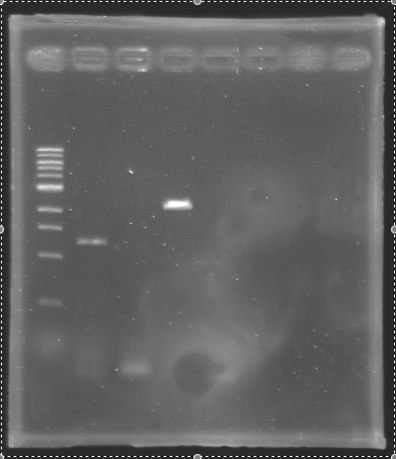
| + | * '''Results :''' | ||
| + | {| border="1" | ||
| + | |- style="text-align: center;" | ||
| + | |'''Name''' | ||
| + | |'''Promotor''' | ||
| + | |align="center"|'''Gel''' | ||
| + | |align="center"|'''Band''' | ||
| + | |align="center"|'''Expected size''' | ||
| + | |align="center"|'''Measured size''' | ||
| + | |- style="text-align: center;" | ||
| + | |PCR_140 | ||
| + | | | ||
| + | |Gel 3 | ||
| + | |2 | ||
| + | |876 bp | ||
| + | |style="background: #cbff7B"|<center> 900 bp </center> | ||
| + | |- style="text-align: center;" | ||
| + | |PCR_141 | ||
| + | |Negative Control | ||
| + | |Gel 3 | ||
| + | |3 | ||
| + | |0 bp | ||
| + | |style="background: #cbff7B"|<center> 0 bp </center> | ||
| + | |- style="text-align: center;" | ||
| + | |PCR_142 | ||
| + | | | ||
| + | |Gel 3 | ||
| + | |4 | ||
| + | |351 bp | ||
| + | |style="background: #cbff7B"|<center> 350 bp </center> | ||
| + | |- style="text-align: center;" | ||
| + | |PCR_143 | ||
| + | |Negative Control | ||
| + | |Gel 3 | ||
| + | |5 | ||
| + | |0 bp | ||
| + | |style="background: #cbff7B"|<center> 0 bp </center> | ||
| + | |- style="text-align: center;" | ||
| + | |PCR_144 | ||
| + | | | ||
| + | |Gel 3 | ||
| + | |6 | ||
| + | |579 bp | ||
| + | |style="background: #cbff7B"|<center> 600 bp </center> | ||
| + | |- style="text-align: center;" | ||
| + | |PCR_145 | ||
| + | |Negative Control | ||
| + | |Gel 3 | ||
| + | |7 | ||
| + | |0 bp | ||
| + | |style="background: #cbff7B"|<center> 0 bp </center> | ||
==Culture of ligation transformants== | ==Culture of ligation transformants== | ||
Revision as of 16:53, 12 August 2008
Transformation
DigestionPCRWe performed PCR on to amplify the sequence in order to have enough amount of DNA to carry out the following of our experiments.
PCR amplificationProtocol
For each sample, 1 µl dNTP
PCR verification/AnalysisAfter the PCR :
ladder : 10µl ladder 1 kb
ladder : 10µl ladder 100 bp
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
 "
"