Team:Paris/August 14
From 2008.igem.org
(Difference between revisions)
(→Results of the PCR of the wonderful promoter of fliL) |
|||
| Line 1: | Line 1: | ||
{{Paris/Calendar_Links|August 13|August 15}} | {{Paris/Calendar_Links|August 13|August 15}} | ||
| - | |||
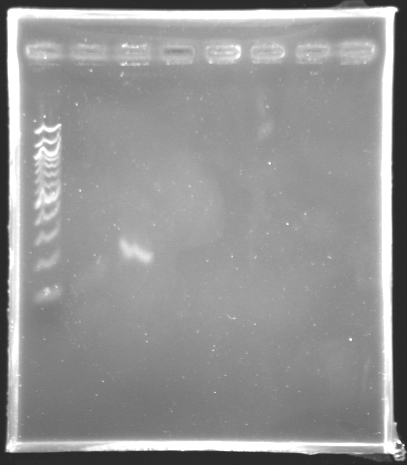
=='''Results of the PCR of the wonderful promoter of ''fliL'' '''== | =='''Results of the PCR of the wonderful promoter of ''fliL'' '''== | ||
[[Image:KR000147.jpg|thumb|Amplification of pfliL]] | [[Image:KR000147.jpg|thumb|Amplification of pfliL]] | ||
| - | |||
==='''Electrophoresis settings :'''=== | ==='''Electrophoresis settings :'''=== | ||
| Line 38: | Line 36: | ||
We observe that the bands are curved, we suppose that the wells were not very clean. | We observe that the bands are curved, we suppose that the wells were not very clean. | ||
The size of fliL is good, we will digest it and ligate it today. | The size of fliL is good, we will digest it and ligate it today. | ||
| + | |||
| + | |||
=='''Digestion and Ligation of the wonderful promoter of ''fliL'' '''== | =='''Digestion and Ligation of the wonderful promoter of ''fliL'' '''== | ||
| Line 98: | Line 98: | ||
====Washing of the digestions==== | ====Washing of the digestions==== | ||
We washed the DNA following the [[Team:Paris/Notebook/Protocols#Purification_.28Kit_Promega.29|standard protocol]]. | We washed the DNA following the [[Team:Paris/Notebook/Protocols#Purification_.28Kit_Promega.29|standard protocol]]. | ||
| + | |||
===Ligation=== | ===Ligation=== | ||
| Line 123: | Line 124: | ||
| 6 | | 6 | ||
|} | |} | ||
| + | |||
| + | |||
==PCR screening of cloning of E0240, flhD, flhC and pflhDC+flhDC== | ==PCR screening of cloning of E0240, flhD, flhC and pflhDC+flhDC== | ||
Latest revision as of 14:58, 16 August 2008
Results of the PCR of the wonderful promoter of fliLElectrophoresis settings :
Electrophoresis Results :
Remarks: We observe that the bands are curved, we suppose that the wells were not very clean. The size of fliL is good, we will digest it and ligate it today.
Digestion and Ligation of the wonderful promoter of fliLAs the primer used to amplify the promoter of fliL had only two nucleotides after the restriction sites, we tried the two digestions possible : EcoRI + SpeI and XbaI + PstI. DigestionProtocol :
Washing of the digestionsWe washed the DNA following the standard protocol.
Ligation
PCR screening of cloning of E0240, flhD, flhC and pflhDC+flhDC
|
 "
"