Team:Paris/August 27
From 2008.igem.org
(Difference between revisions)
(→Ligation) |
(→Measurement of the concentration of D187 purified) |
||
| Line 13: | Line 13: | ||
[[Team:Paris/Notebook/Protocols#Concentration_of_the_Miniprep | Protocol (it's same that for Miniprep)]] | [[Team:Paris/Notebook/Protocols#Concentration_of_the_Miniprep | Protocol (it's same that for Miniprep)]] | ||
| - | + | => the experiments have failed, so we need to repeat the step of digestion. | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
=Cloning of EnvZ* in pSB1A2= | =Cloning of EnvZ* in pSB1A2= | ||
Revision as of 18:30, 27 August 2008
|
Construction of pFlhB - mRFP Tripart (LVA+)Aim : Construction of "pFlhB-RBS-mRFP-dbl ter" (pFlhB-I732078)
DigestionMeasurement of the concentration of D187 purifiedProtocol (it's same that for Miniprep) => the experiments have failed, so we need to repeat the step of digestion. Cloning of EnvZ* in pSB1A2Transformation results
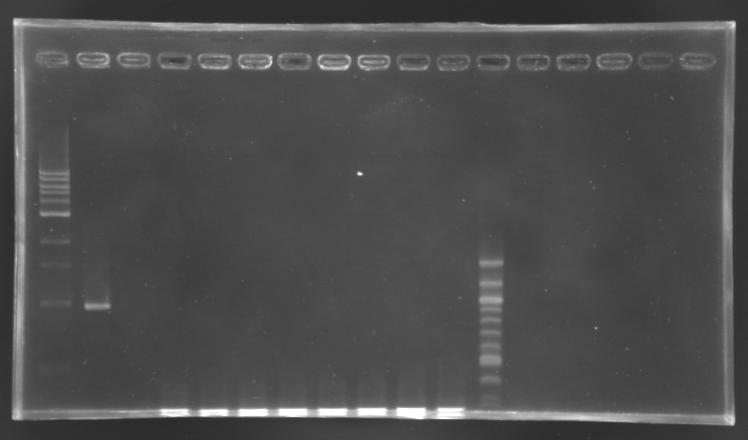
PCR screening
Electrophoresis
PCR
elongation time: 2 min 30
Cloning of OmpR*DigestionDetermination of the concentration of DNAWe used the biophotometer
Name of the digestions
Protocol of digestion
Cleaning of the digestion productsLigationDetermination of the concentration of DNAWe used the biophotometer
Protocol of ligation L171
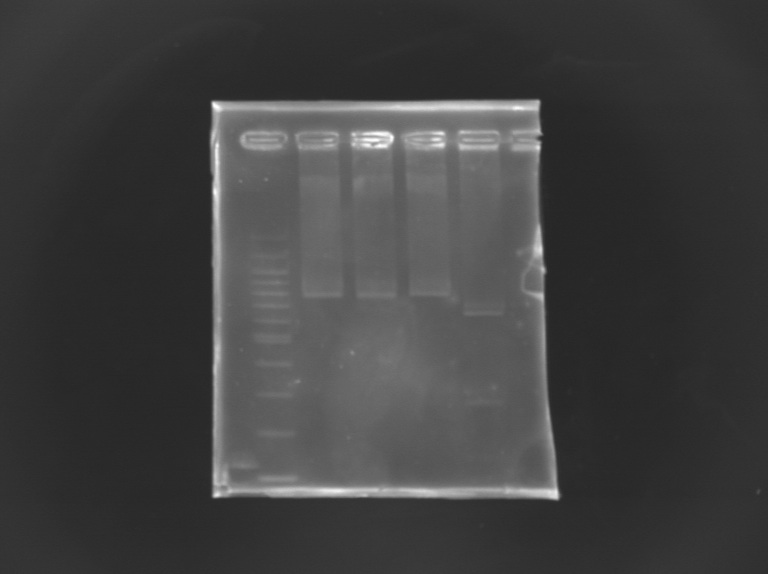
Checking mutagenesis FliAFor this, i digested mutated FliA and non-mutated FliA with EcoRI and PstI and put in migration the digestion products running on gels. |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
 "
"