|
← Yesterday ↓ Calendar ↑Tomorrow →
Construction of pFlhB - mRFP Tripart (LVA+)
Aim : Construction of "pFlhB-RBS-mRFP-dbl ter" (pFlhB-I732078)     
We can only do the construction with mRFP Tripart (LVA+) because the stable strain with the Biobricks I732011 (mRFP Tripart LVA-) don't to growth.
Digestion
Measurement of the concentration of D187 purified
Protocol (it's same that for Miniprep)
=> the experiments of Gel Extraction have failed, so we need to repeat the step of digestion.
Digestion
Protocol Digestion
| Name
| Template DNA
| Description
| Vol MP (µl)
| Vol H2O (µl)
| Enzymes
|
| D187
| MP168.1
| RBS - mRFP - term (FV)
| 9.00
| 15.7
| EcoRI and XbaI
|
Gel Verification
Protocol
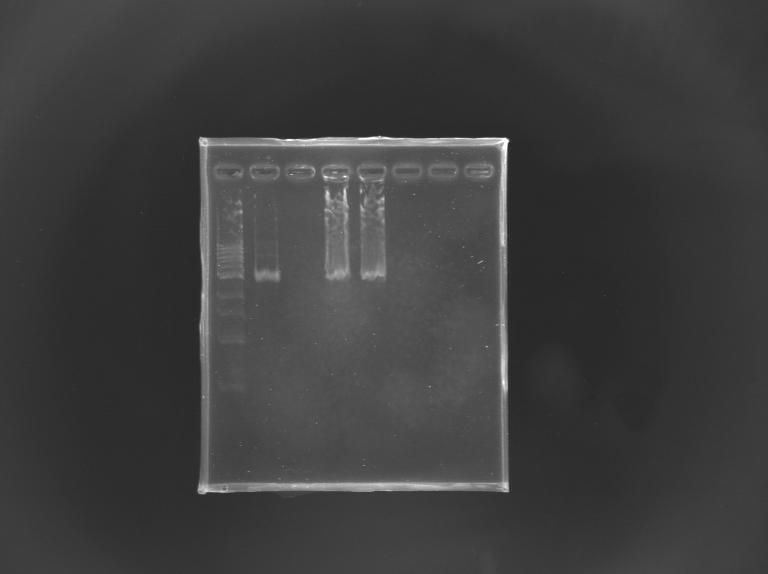 Gel Verification of D187 digestion
| Well
| 1
| 2
| 3
| 4
| 5
| 6
|
| Sample
| 100 pb ladder
| MP168.1
| no sample
| D187
| no sample
|
| Expected size (pb)
|
| 2 955
|
| 2 940
|
|
| Measured size (pb)
|
| 2 900
|
| 2 900
|
|
Cloning of EnvZ* in pSB1A2
Transformation results
|
| control
| insert / vector mass ratio
|
| transformation with pUC19
| transformation without plasmid
| ligation without insert
| 1,8 / 1
| 2,4 / 1
| 3,1 / 1
|
| number of colonies
| many
| 0
| 0
| 9
| 6
| 2
|
| number of clones picked up for screening
|
| 4
| 2
| 2
|
| number of colonies picked up for screening bis
|
| 4
| 4
|
|
PCR screening
- screening programm
- elongation time: 2 min
- number of cycle: 24
- total volume reaction: 25 µL
- primers used: O18 and O19
- positive control: S158 (pSB3K3)
- negative control: no template
Electrophoresis
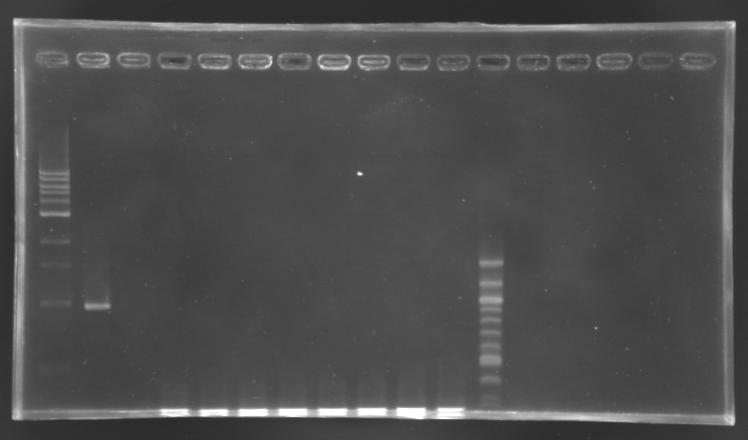 Screening of the cloning of EnvZ* - clones 1 to 8
| well n°
| 1
| 2
| 3
| 4
| 5
| 6
| 7
| 8
| 9
| 10
| 11
| 12
|
| sample
| 1 kb DNA ladder
| positive
control
| negative
control
| L165.1
| L165.2
| L165.3
| L165.4
| L165.5
| L165.6
| L165.7
| L165.8
| 100 bp DNA ladder
|
| expected size
|
|
|
| 1659 bp
|
|
| measured size
|
|
|
| 0,3 kb
|
|
No correct clone
The 8 other clones were also screened.
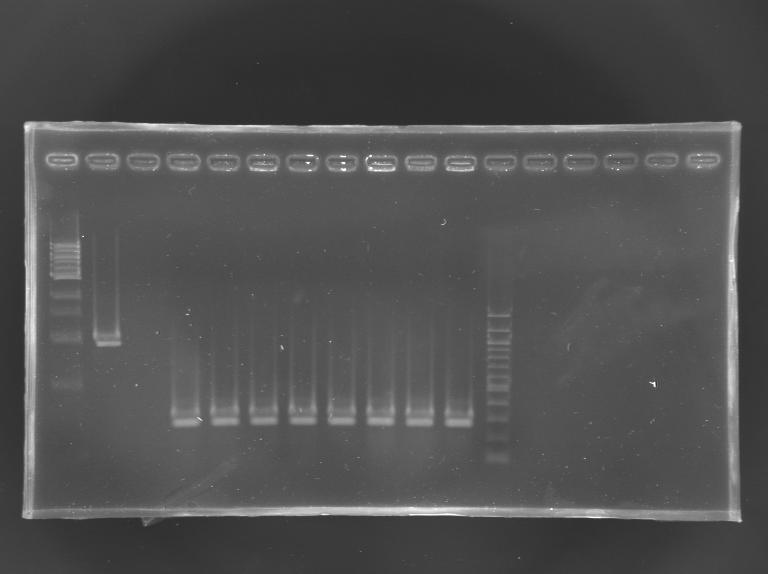 Screening of the cloning of EnvZ* - clones 9 to 16 PCR
elongation time: 2 min 30
Electrophoresis
| well n°
| 1
| 2
| 3
| 4
| 5
| 6
| 7
| 8
| 9
| 10
| 11
| 12
|
| sample
| 1 kb DNA ladder
| positive
control
| negative
control
| L165.9
| L165.10
| L165.11
| L165.12
| L165.13
| L165.14
| L165.15
| L165.16
| 100 bp DNA ladder
|
| expected size
|
|
|
| 1659 bp
|
|
| measured size
|
|
|
| 0,3 kb
|
|
Results: All the clones analysed were not correct.
Cloning of OmpR*
Digestion
Determination of the concentration of DNA
We used the biophotometer
- 5 µL of template DNA or 5 µL of EB buffer for th blank
- 55 µL of pure water
| Template DNA
| Concentration of DNA
|
| PCR 147
| 150 µg/mL
|
| PCR 148
| 101 µg/mL
|
| MP 101.2
| 353 µg/mL
|
Name of the digestions
| Name of the digestion
| Template DNA
| What's in?
| Enzymes used
|
| D 188
| PCR 148
| OmpR*
| XbaI-PstI
|
| D 189
| MP 101.2
| pSB1A2
| XbaI-PstI
|
Protocol of digestion
- D 188 : 3 µL of PCR 148
- D 189 : 3 µL of MP 101.2
- 3µL Buffer (n°2) 10X
- 0.3µL BSA 100X
- 22.7 µL of pure Water
- 1 µL of each enzyme
- Incubate during about 2h30 at 37°C
- 20 minutes at 65°C
Cleaning of the digestion products
Standard protocol.
Ligation
Determination of the concentration of DNA
We used the biophotometer
- 10 µL of template DNA or 10 µL of EB buffer for th blank
- 50 µL of pure water
| Template DNA
| Concentration of DNA
|
| D 188
| 9 µg/mL
|
| D 189
| 30 µg/mL
|
Protocol of ligation L171
- 2 µL Ligase Buffer 10X
- 1.5 µL D 189 (vector)
- 5 µL D 188 (insert)
- 11.5 µL pure Water (qsp 20 µL)
- 1 µL T4 DNA ligase at 400 000 U/mL concentration
- O/N at 16°C
Checking mutagenesis FliA
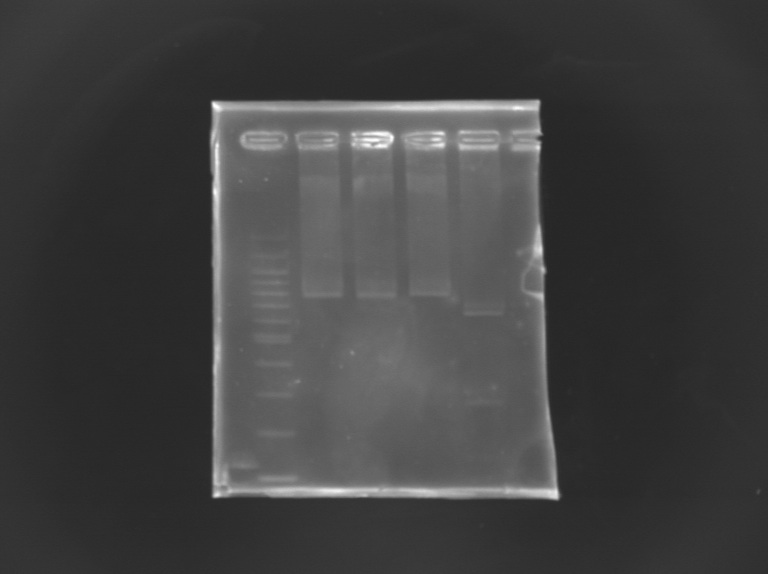 EcorI/PstI digestion of mutated FliA For this, i digested mutated FliA and non-mutated FliA with EcoRI and PstI and put in migration the digestion products running on gels.
Results : No digestion for the mutated sequence --> successful mission !
|
 "
"




