Team:Paris/August 7
From 2008.igem.org
(→Results of the PCR we did last night) |
(→Preparation of the newly ammplified promoters) |
||
| (35 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
{{Paris/Calendar_Links|August 6|August 8}} | {{Paris/Calendar_Links|August 6|August 8}} | ||
| + | |||
| + | ==Glycerol Stocks== | ||
| + | *1mL of each culture (with 2 clones) has been added to 1mL of 40% glycerol. | ||
| + | *For each clone, two glycerol stocks have been done. | ||
| + | *Stored at -20°C. | ||
| + | |||
| + | {|border="1" | ||
| + | |'''name''' | ||
| + | |'''Strain''' | ||
| + | |- | ||
| + | |S141 | ||
| + | |MG1655 | ||
| + | |- | ||
| + | |S142 | ||
| + | |J61002 | ||
| + | |} | ||
| + | |||
| + | |||
==Result of the isolation of colonies== | ==Result of the isolation of colonies== | ||
===E0240 and pSB3K3=== | ===E0240 and pSB3K3=== | ||
| Line 8: | Line 26: | ||
<br> We checked in the strain library, actually the strains do not carry any resistance cassette. We plated them once again on petri dishes with LB without antibiotics | <br> We checked in the strain library, actually the strains do not carry any resistance cassette. We plated them once again on petri dishes with LB without antibiotics | ||
| - | == | + | ==Preparation of the newly amplified promoters== |
| + | ===Electrophoresis of the PCR products made [[Team:Paris/August_6|yesterday]]=== | ||
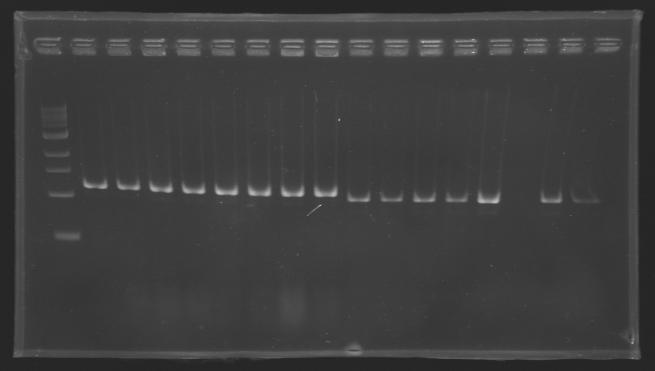
[[image:KR000116.jpg|thumb|Standart PCR to amplify pflgA, pflgB and pflhB(Gel1)]] | [[image:KR000116.jpg|thumb|Standart PCR to amplify pflgA, pflgB and pflhB(Gel1)]] | ||
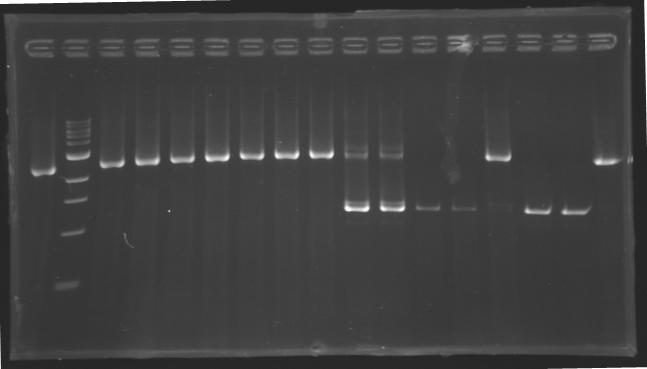
[[image:KR000117.jpg|thumb|PCR with gradient to amplify pflhDC(Gel2)]] | [[image:KR000117.jpg|thumb|PCR with gradient to amplify pflhDC(Gel2)]] | ||
| Line 14: | Line 33: | ||
* Gel : 1.5 % agar | * Gel : 1.5 % agar | ||
* 3µL template DNA | * 3µL template DNA | ||
| - | * QuickLoad DNA ladder 100 bp | + | * 10µL QuickLoad DNA ladder 100 bp |
{|- align = "center" | border="1" | {|- align = "center" | border="1" | ||
| Line 54: | Line 73: | ||
<br><br><br> | <br><br><br> | ||
'''Results''' | '''Results''' | ||
| - | *We have no results for pflhDC, wo don't know yet where is the problem. We will try with other conditions! (yet undetermined) | + | *We have '''no results for pflhDC''', wo don't know yet where is the problem. We will try with other conditions! (yet undetermined) |
| - | *Concerning pflhB, pflgA and pflgB, the protocol seems to be very operational: we always have great results ! | + | *Concerning '''pflhB, pflgA and pflgB''', the protocol seems to be '''very operational''': we always have great results ! |
| + | |||
| + | ===Washing of the PCR products=== | ||
| + | *Kit used : Wizard SV Gel and PCR Clean-Up System from Promega | ||
| + | *Standart protocol except the last step. Instead of eluting with water, we used 30 µL of BE buffer (from Qiagen) | ||
| + | |||
| + | ===DNA concentration measurement=== | ||
| + | We used two methods: | ||
| + | ====With a Spectrophotometer==== | ||
| + | *λ = 260 nm | ||
| + | *White: 100µL BE Buffer | ||
| + | *Templates : 2 µL DNA + 98 µL BE Buffer | ||
| + | |||
| + | |||
| + | {| Border="1" | ||
| + | |align="center"|'''Template''' | ||
| + | |align="center"|'''Absorbance''' | ||
| + | |align="center"|'''Estimated'''<br>'''Concentration '''<br>(µg/µL) | ||
| + | |- | ||
| + | |align="center"|pflgA | ||
| + | |align="center"|0.202 | ||
| + | |align="center"|0.5 | ||
| + | |- | ||
| + | |align="center"|pflgB | ||
| + | |align="center"|0.210 | ||
| + | |align="center"|0.5 | ||
| + | |- | ||
| + | |align="center"|pflhA | ||
| + | |align="center"|0.193 | ||
| + | |align="center"|0.5 | ||
| + | |} | ||
| + | |||
| + | ====With a Biophotometer==== | ||
| + | {| Border="2" | ||
| + | |align="center"|'''Template''' | ||
| + | |align="center"|'''Estimated'''<br>'''Concentration '''<br>(µg/µL) | ||
| + | |align="center"|'''Ratio DO260/DO280''' | ||
| + | |- | ||
| + | |align="center"|pflgA | ||
| + | |align="center"|0.05 | ||
| + | |align="center"|1.06 | ||
| + | |- | ||
| + | |align="center"|pflgB | ||
| + | |align="center"|0.1 | ||
| + | |align="center"|1.44 | ||
| + | |- | ||
| + | |align="center"|pflhA | ||
| + | |align="center"|0.O5 | ||
| + | |align="center"|1.66 | ||
| + | |} | ||
| + | Remarks : | ||
| + | * There is a great difference between the two methods : sometimes 1 log ! | ||
| + | * The electrophoresis of the PCR products showed that pflgB was more concentrated than pflgA and pflhA. As a consequence, we rely on the figures determined by the Biophotometer | ||
| + | |||
| + | ===Digestion=== | ||
| + | |||
| + | ====Protocol==== | ||
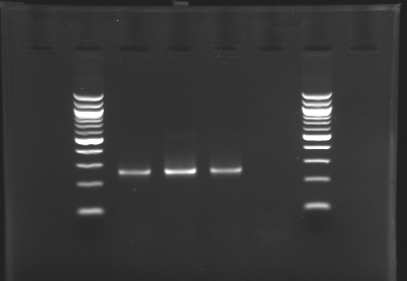
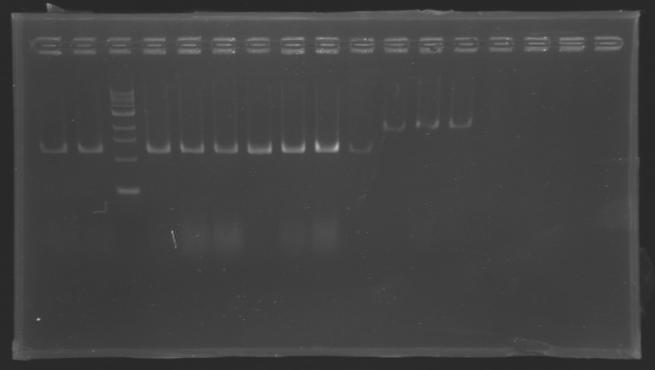
| + | [[Image:KR000126.jpg|thumb|Gel 1 - 1% agar, ladder 1kb]] | ||
| + | [[Image:KR000127.jpg|thumb|Gel 2 - 1.5 % agar, ladder 100bp]] | ||
| + | {| Border="2" | ||
| + | |align="center"|'''Digestion name''' | ||
| + | |align="center"|'''Template DNA''' | ||
| + | |align="center"|'''Enzymes''' | ||
| + | |align="center"|'''Quantity of DNA used''' | ||
| + | |- | ||
| + | |align="center"|D132 | ||
| + | |align="center"|PCR 124 - pflgA | ||
| + | |align="center"|EcoRI-SpeI | ||
| + | |align="center"|2 µL | ||
| + | |- | ||
| + | |align="center"|D133 | ||
| + | |align="center"|PCR 125 - pflgB | ||
| + | |align="center"|EcoRI-SpeI | ||
| + | |align="center"|1 µL | ||
| + | |- | ||
| + | |align="center"|D134 | ||
| + | |align="center"|PCR 16 - pflhB | ||
| + | |align="center"|EcoRI-SpeI | ||
| + | |align="center"|2 µL | ||
| + | |- | ||
| + | |align="center"|D136 | ||
| + | |align="center"|MP123 | ||
| + | |align="center"|EcoRI-SpeI | ||
| + | |align="center"|8 µL | ||
| + | |} | ||
| + | '''Digestion mix:''' | ||
| + | *X µL of template DNA | ||
| + | *1 µL Enzyme 1 | ||
| + | *1 µL Enzyme 2 | ||
| + | *3 µL Buffer P2 | ||
| + | *0.3 µL BSA | ||
| + | *24.7 - X µL of distilled water | ||
| + | |||
| + | We pu the digestion mix to incubate during 2h at 37°C | ||
| + | |||
| + | ====Results==== | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | {|- align = "center" | border="1" | ||
| + | |align="center"|'''Name''' | ||
| + | |align="center"|'''Gel''' | ||
| + | |align="center"|'''Band''' | ||
| + | |align="center"|'''Expected size''' | ||
| + | |align="center"|'''Measured size''' | ||
| + | |- | ||
| + | |align="center"|D136 | ||
| + | |align="center"|1 | ||
| + | |align="center"|2 | ||
| + | |style="background: #cbff7B"|<center>2925 pb</center> | ||
| + | |align="center"|3000 pb | ||
| + | |- | ||
| + | |align="center"|MP 123 | ||
| + | |align="center"|1 | ||
| + | |align="center"|4 | ||
| + | |style="background: #cbff7B"|<center>3002 pb</center> | ||
| + | |align="center"|3000 pb | ||
| + | |- | ||
| + | |align="center"|D133 | ||
| + | |align="center"|2 | ||
| + | |align="center"|2 | ||
| + | |style="background: #ff6d73"|<center>225 pb</center> | ||
| + | |align="center"|nothing | ||
| + | |- | ||
| + | |align="center"|D134 | ||
| + | |align="center"|2 | ||
| + | |align="center"|3 | ||
| + | |style="background: #ff6d73"|<center>225 pb</center> | ||
| + | |align="center"|nothing | ||
| + | |- | ||
| + | |align="center"|D135 | ||
| + | |align="center"|2 | ||
| + | |align="center"|4 | ||
| + | |style="background: #ff6d73"|<center>226 pb</center> | ||
| + | |align="center"|nothing | ||
| + | |} | ||
| + | |||
| + | |||
| + | Conclusion : The digestion of MP123 into D136 worked very well. [[Team:Paris/August_8|Tomorrow]] we will isolate it. | ||
| + | We see nothing on the other digestion. Maybe [https://2008.igem.org/Image:Cyprien-Maisonnier.jpg someone] forgot to put the template DNA inside the tube !? | ||
| + | Anyway, we will do the manipulation again [[Team:Paris/August_8|tomorrow]]. | ||
==Transformations== | ==Transformations== | ||
===Protocol=== | ===Protocol=== | ||
| - | Use of TOP10 chemically | + | Use of TOP10 chemically competent cells |
* Defroze competent cells on ice during 5' | * Defroze competent cells on ice during 5' | ||
| - | * Add 5µl of | + | * Add 5µl of Ligation products in 50µL of competent bacteria (or 1µL for the positive control puc19) |
* Incubate 30' on ice | * Incubate 30' on ice | ||
* Heat-shock the cells during 30" at 42°C without shaking | * Heat-shock the cells during 30" at 42°C without shaking | ||
| Line 70: | Line 230: | ||
* Spin at 5.000rpm during 30" | * Spin at 5.000rpm during 30" | ||
* Remove 150µL of supernatant | * Remove 150µL of supernatant | ||
| - | * | + | * Resuspend the pellet in the 150µL left |
| - | * Spread on | + | * Spread on adequate plates |
* Incubate O/N at 37°C | * Incubate O/N at 37°C | ||
| - | |||
=== List of the Ligation Transformation === | === List of the Ligation Transformation === | ||
| Line 161: | Line 320: | ||
===Results=== | ===Results=== | ||
| + | |||
| + | |||
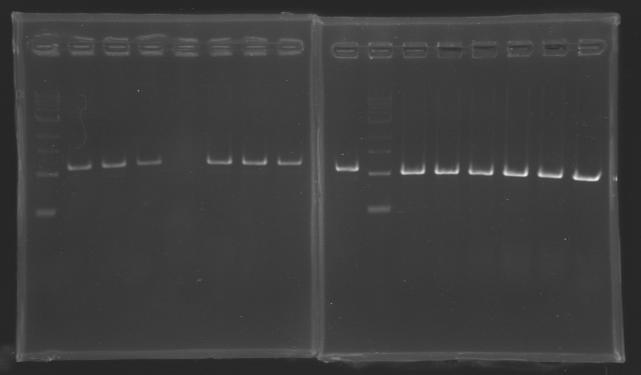
| + | Gel 1 : L100-L101[[Image: KR000118_gel1.jpg|200px]] | ||
| + | Gel 2 : L113-L114[[Image: KR000119_gel2.jpg|200px]] | ||
| + | Gel 3 : L120-L122[[Image: KR000120_gel3.jpg|200px]]<br> | ||
| + | Gel 4 : L123-L126[[Image: KR000122_gel4.jpg|200px]] | ||
| + | Gel 5 : L126[[Image: KR000123_gel5.jpg|100px]] | ||
* | * | ||
{| border="1" | {| border="1" | ||
| - | + | ||
| - | + | |align="center"|'''name''' | |
| - | + | |align="center"|'''Description''' | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | |align="center"|''' | + | |
| - | |align="center"|''' | + | |
|align="center"|'''Expected size''' | |align="center"|'''Expected size''' | ||
|align="center"|'''Measured size''' | |align="center"|'''Measured size''' | ||
| + | |align="center"|'''Gel''' | ||
|align="center"|'''Band''' | |align="center"|'''Band''' | ||
| + | |align="center"|'''Comments''' | ||
|- | |- | ||
| - | |align="center"| | + | |align="center"|PCR1_’’’L101(1-8)’’’ |
| - | |align="center"| | + | |align="center"|D110-D131 |
| + | |align="center"|1200 | ||
| + | |align="center"|1100 | ||
| + | |align="center"|1 | ||
|align="center"|2-9 | |align="center"|2-9 | ||
| - | |align="center"| | + | |align="center"|(1-8) ok |
| - | |align="center"| | + | |- |
| + | |align="center"|PCR2_’’’L102(1-8)’’’ | ||
| + | |align="center"|D129-D118 | ||
| + | |align="center"|1045 | ||
| + | |align="center"|1000 | ||
| + | |align="center"|1 | ||
|align="center"|10-17 | |align="center"|10-17 | ||
| - | |align="center"| | + | |align="center"|(1-8) ok |
| - | |align="center"| | + | |- |
| + | |align="center"|PCR3_’’’L113(1-8)’’’ | ||
| + | |align="center"|D126-D130 | ||
| + | |align="center"|1239 | ||
| + | |align="center"|2500 | ||
| + | |align="center"|2 | ||
|align="center"|1, 3-9 | |align="center"|1, 3-9 | ||
| - | |align="center"| | + | |align="center"|to do again |
| - | |align="center"| | + | |- |
| + | |align="center"|PCR4_’’’L114(1-8)’’’ | ||
| + | |align="center"|D126-D131 | ||
| + | |align="center"|1239 | ||
| + | |align="center"|1200 | ||
| + | |align="center"|2 | ||
|align="center"|10-17 | |align="center"|10-17 | ||
| + | |align="center"|(1-8) ok | ||
|- | |- | ||
| - | | | + | |align="center"|PCR5_’’’L120(1-8)’’’ |
| - | + | |align="center"|D106-D130 | |
| - | + | |align="center"|1239 | |
| - | + | |align="center"|2000 | |
| - | + | |align="center"|3 | |
| - | + | |align="center"|1,2, 4-9 | |
| - | + | |align="center"|(1-8) ok | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | |align="center"| | + | |
| - | |align="center"| | + | |
| - | |align="center"| | + | |
| - | |align="center"| | + | |
| - | |align="center"| | + | |
| - | |align="center"| | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
|- | |- | ||
| - | |align="center"| | + | |align="center"|PCR6_’’’L122(1-4)’’’ |
| - | |align="center"| | + | |align="center"|D107-D130 |
| - | |align="center"| | + | |align="center"|1239 |
| - | |align="center"| | + | |align="center"|1200 |
| - | |align="center"| | + | |align="center"|3 |
|align="center"|10-13 | |align="center"|10-13 | ||
| - | |align="center"| | + | |align="center"|(1) ok |
| - | |align="center"| | + | |- |
| + | |align="center"|PCR7_’’’L123(1-8)’’’ | ||
| + | |align="center"|D107-D131 | ||
| + | |align="center"|1200 | ||
| + | |align="center"|1100 | ||
| + | |align="center"|4 & 4' | ||
|align="center"|2-8, 1 | |align="center"|2-8, 1 | ||
| - | |align="center"| | + | |align="center"|(1-8) ok |
| - | |align="center"| | + | |- |
| + | |align="center"|PCR8_’’’L126(1-6)’’’ | ||
| + | |rowspan="2"|D102-D118 | ||
| + | |rowspan="2"|1045 | ||
| + | |align="center"|1000 | ||
| + | |align="center"|4' | ||
|align="center"|3-8 | |align="center"|3-8 | ||
| + | |rowspan="2"|(1-8) ok | ||
|- | |- | ||
| - | | | + | |align="center"|PCR5_’’’L126(7-8)’’’ |
| - | + | |align="center"|1000 | |
| - | + | |align="center"|6 | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | |align="center"| | + | |
| - | |align="center"| | + | |
|align="center"|2-3 | |align="center"|2-3 | ||
| - | |||
| - | |||
|} | |} | ||
| - | ==> '''Conclusion :''' | + | ==> '''Conclusion :'''In futur , we will prepare minipreps and stocks for the transformant clones succesfull. |
Latest revision as of 12:42, 13 August 2008
Glycerol Stocks
Result of the isolation of coloniesE0240 and pSB3K3E0240 and pSB3K3 are ok : there is a lot of single colonies S120 and S121S120 and S121 : there is a problem, there is nothing on the plates. We have to check whether those strains are really resistant to Amp.
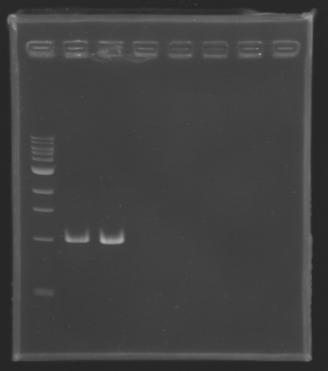
Preparation of the newly amplified promotersElectrophoresis of the PCR products made yesterdayElectrophoresis settings
Washing of the PCR products
DNA concentration measurementWe used two methods: With a Spectrophotometer
With a Biophotometer
Remarks :
DigestionProtocol
Digestion mix:
We pu the digestion mix to incubate during 2h at 37°C Results
Conclusion : The digestion of MP123 into D136 worked very well. Tomorrow we will isolate it. We see nothing on the other digestion. Maybe someone forgot to put the template DNA inside the tube !? Anyway, we will do the manipulation again tomorrow. TransformationsProtocolUse of TOP10 chemically competent cells
List of the Ligation Transformation
PCR Screening of Ligation Transformants of 1st AugustUse of 8 clones of Ligation transformants for screening PCR
Protocol of screening PCR
Conditions of electrophoresis
ResultsGel 1 : L100-L101
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
 "
"