|
← Yesterday ↓ Calendar ↑Tomorrow →
Extraction of pSB3K3 et E0240 in pSB1A2 plasmid from overnight bacteria culture using the QIAspin Miniprep Kit (QIAGEN) by QIACube.
- Carried out 2 times (2 tubes)
| name
| Biobrick
| plasmid
|
| MP142
| -
| pSB3K3
|
| MP143
| E0240
| pSB1A2
|
Amplification of Genes of interest (OmpR, EnvZ, FlhDC)
We performed PCR on to amplify the sequence in order to have enough amount of DNA to carry out the following of our experiments.
PCR amplification
Protocol
| Number
| Name
| Sequence
| Length
| Comments
|
| O126
| Gene-EnvZ-F
| GTTTCTTCGAATTCGCGGCCGCTTCTAGATGAGGCGATTGCGCTTCTCGCCAC
| 53
|
|
| O127
| Gene-EnvZ-R
| GTTTCTTCGAATTCGCGGCCGCTTCTAGTTATTACCCTTCTTTTGTCGTGCCCTGCGCC
| 59
|
|
| O131
| Gene-FlhC-R
| GTTTCTTCGAATTCGCGGCCGCTTCTAGTTATTAAACAGCCTGTACTCTCTGTTCATCC
| 59
|
|
| O132
| Gene-FlhD-F
| GTTTCTTCGAATTCGCGGCCGCTTCTAGATGCATACCTCCGAGTTGCTGAAAC
| 53
| Don't amplify the natural rbs of FlhD
|
| O138
| Gene-OmpR-F
| GTTTCTTCGAATTCGCGGCCGCTTCTAGATGCAAGAGAACTACAAGATTCTGG
| 53
|
|
| O139
| Gene-OmpR-R
| GTTTCTTCGAATTCGCGGCCGCTTCTAGTTATTATGCTTTAGAGCCGTCCGGTACAAAG
| 59
|
|
- Preparation of the templates :
Resuspension of 1 colony in 100µl of water.
For each samples,
1 µl dNTP
10 µl Buffer Phusion 5x
2,5 µl Oligo_F
2,5 µl Oligo_R
1µl template
1 µl Phusion
50 µl qsp H2O (33µl)
| Name
| genes
| Oligo
| templates
|
| PCR_127
| FlhDC
| O131_O132
| MG1655
|
| PCR_128
| OmpR
| O138_O139
| Strain OmpR*
|
| PCR_129
| EnvZ
| O126_O127
| Strain EnvZ*
|
| PCR_Control -
| -
| O126_O127
| Water
|
- Program PCR_Screening : Annealing 55°C - Time élongation 1'30" - Number cycle : 29
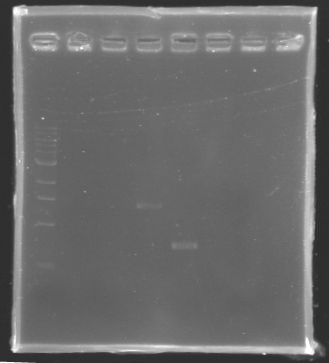
PCR verification/Analysis
After the PCR :
- 3µl have been analysed by electrophoresis
- the other 47µl of PCR products have been purified by the Promega kit.
ladder : 10µl ladder 1 kb
samples : 3µl of PCR products + 2µl of Loading Dye
migration 30min at 100W, on a 1% agarose gel
| Name
| Promotor
| Band
| Expected size
| Measured size
|
| PCR_Control -
| control -
| 2
| 0 pb
| 0 pb
|
| PCR_127
| FlhDC gene
| 3
| 972 pb
| 0 pb
|
| PCR_128
| OmpR gene
| 5
| 762 pb
| 700 pb
|
| PCR_129
| EnvZ gene
| 4
| 1421 pb
| 1400 pb
|
==> Conclusion : we observed the size expected for the PCR products, but not for FlhDC gene.
We hypothesis that its value of amplification for this gene it's low as we can't visualize it. So we try to continue to experiments to know if there is something inside or not this tube.
- Quantification of the PCR products purified
Blank : 2µl of buffer EB + 98µl of water
Samples : 2µl of PCR purified + 98µl of water.
| Name
| Genes
| C° (µg/ml)
| DO 260/280
|
| PCR_127
| FlhDC
| 350
| 1.68
|
| PCR_128
| OmpR
| 150
| 1.97
|
| PCR_129
| EnvZ
| 100
| 1.77
|
| MP 108 cl2 (23 july)
| -vector-
| 150
| 1.84
|
Digestion of PCR products
Protocol :
| Name
| Genes
| Water
| DNA
| Buffer n°2 10X
| BSA 100X
| EcoRI
| PstI
|
| D139
| FlhDC
| 23.7µl
| 1µl
| 3.0µl
| 0.30µl
| 1µl
| 1µl
|
| D140
| OmpR
| 22.7µl
| 2µl
| 3.0µl
| 0.30µl
| 1µl
| 1µl
|
| D141
| EnvZ
| 21.2µl
| 3.5µl
| 3.0µl
| 0.30µl
| 1µl
| 1µl
|
| D142
| -vector-
| 22.7µl
| 2µl
| 3.0µl
| 0.30µl
| 1µl
| 1µl
|
Analysis by electrophoresis
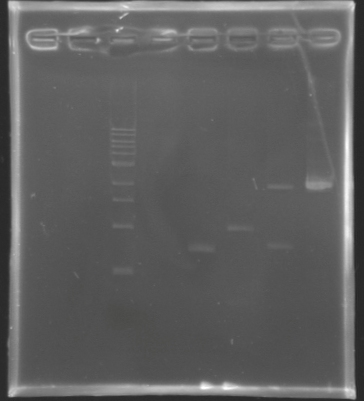 Analysis of PCR product digestion ladder : 10µl ladder 1 kb
samples : 3µl of insert + 2µl of Loading Dye
migration 30min at 100W, on a 1% agarose gel.
| Name
| Promotor
| Band
| Expected size
| Measured size
|
| D139
| FlhDC gene
| 4
| 972 pb
| 0 pb
|
| D140
| OmpR gene
| 5
| 762 pb
| 700 pb
|
| D141
| EnvZ gene
| 6
| 1421 pb
| 1000 pb
|
| D142
| -vector digested-
| 7
| 2057 & 707 pb
| 2000 & 700 pb
|
| D142
| -vector not digested-
| 8
| 2764 pb
| 2100 pb
|
==> Conclusions : We validate the digestion of the vector and the insert. Now we can be sure, that we detect anything for FlhDC genes.
Transformation with promotors of interest
Results
| Name
| Description
| Antibio
| Number of colonies
| Number of red fluorescent colonies
|
| Ligation
|
| L128
| J61002-pFlgA
D136 (FV) - D132 (FI)
| Amp
| ~ 400
| 2
|
| L129
| J61002-pFlgB
D136 (FV) - D133 (FI)
| Amp
| 39
| 5
|
| L130
| J61002-pFlhB
D136 (FV) - D134 (FI)
| Amp
| ~ 1000
| 4 (but 3 are on the edge of the petri dishe)
|
| L131
| J61002-pFlhDC
D136 (FV) - D135 (FI)
| Amp
| 39
| 38
|
| Control
|
| Control 1
| D136
| Amp
| 0
| 0
|
| Positive control
| pUC19
| Amp
| 36
| 0
|
PCR Screening of Ligation Transformants
Use of 8 clones of Ligation transformants for PCR screening
| Ligation
| L128
| L129
| L130
| L131
|
| Name
| pFlgA
| pFlgB
| pFlhB
| pFlhDC
|
| n° clone
| 1
| 2
| 3
| 4
| 5
| 6
| 7
| 8
| 1
| 2
| 3
| 4
| 5
| 6
| 7
| 8
| 1
| 2
| 3
| 4
| 5
| 6
| 7
| 8
| 1
| 2
| 3
| 4
| 5
| 6
| 7
| 8
|
| fluorescence
| red
| red
| no
| no
| no
| no
| no
| no
| red
| red
| red
| red
| red
| no
| no
| no
| red
| no
| no
| no
| no
| no
| no
| no
| red
| red
| red
| red
| red
| red
| red
| no
|
Protocol of screening PCR
25µl of Quick Load 2X
1µl of forward primer 10µM
1µl of reverse primer 10µM
23µl of water
| Ligation
| Name
| primers
|
| L128
| pFlgA
| O100 & O101
|
| L129
| pFlgB
| O102 & O 103
|
| L130
| pFlhB
| O108 & O 109
|
| L131
| pFlhDC
| O111 & O 113
|
- 50µl of PCR Mix by tube/clone
- one toothpick of each clone's colony per tube
- Program : Annealing 55°C - Time élongation 1'30" - Number cycle : 29
- Conditions of electrophoresis
10µl of ladder 100 pb
10µl of screening PCR
migration ~30min at 100W on 1,5% gel
Results of electrophoresis
gel 1 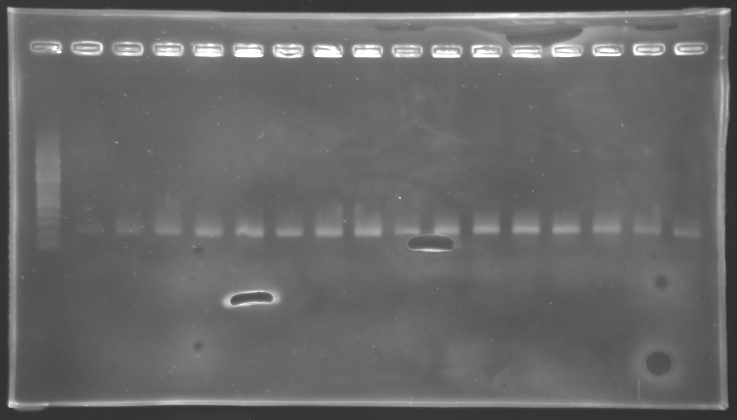 gel 2
gel 2 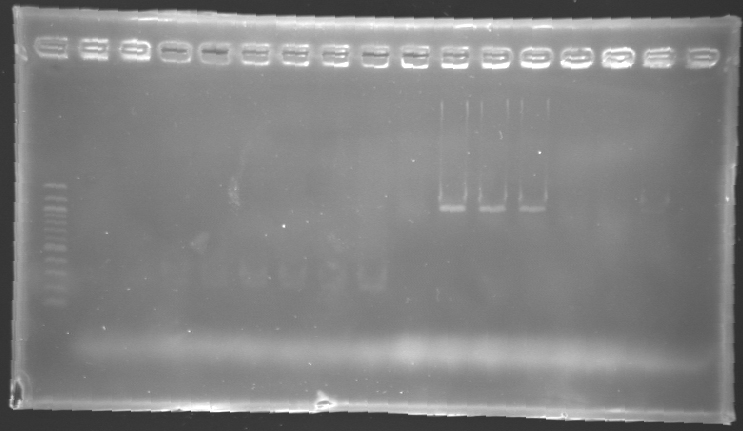
| Name
| Promotor
| Gel
| Band
| Expected size
| Measured size
|
| PCR_124'
| pFlgA
| 1
| 2 to 9
| 261 pb
| 300 pb
|
| PCR_125'
| pFlgB
| 1
| 10 to 17
| 261 pb
| 300 pb
|
| PCR_126'
| pFlhB
| 2
| 2 to 9
| 260 pb
| 300 pb
|
| PCR_127'
| pFlhDC
| 2
| 10 to 17
| 446 pb
| 1,000 pb
|
==> Conclusion:
- PCR of pFlgA, pFlgB and pFlhB have succeed, but we always have a problem with pFlhDC probably because of the primers which are not specific.
|
 "
"