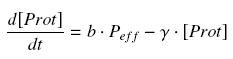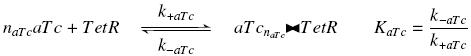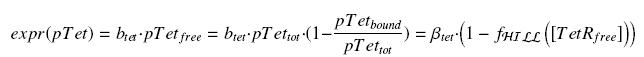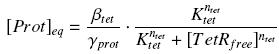Team:Paris/Modeling/estimation
From 2008.igem.org
(→first hypothesis) |
(→By aTc / TetR / Ptet) |
||
| Line 46: | Line 46: | ||
====<center>By aTc / TetR / ''Ptet'' </center>==== | ====<center>By aTc / TetR / ''Ptet'' </center>==== | ||
| - | Now, we must use as a variable of reference an element that could be introduced in the bacteria, well-controlled, and from which all the concentrations of our transcription factor will depend. We propose a construction in which our transcription factor is put after the promoter ''Ptet'', which is under the repression of TetR. Since aTc is a small diffusive molecule that binds to TetR and inhibits this way the repression of ''Ptet'', we can use it as an 'inducer'. To do so, we must place in the bacterium the gene ''tetR'' after a constitutive promoter (like J23101). According to previous hypothesis, this will provide at steady-state a 'constant concentration' of TetR (we note [TetR | + | Now, we must use as a variable of reference an element that could be introduced in the bacteria, well-controlled, and from which all the concentrations of our transcription factor will depend. We propose a construction in which our transcription factor is put after the promoter ''Ptet'', which is under the repression of TetR. Since aTc is a small diffusive molecule that binds to TetR and inhibits this way the repression of ''Ptet'', we can use it as an 'inducer'. To do so, we must place in the bacterium the gene ''tetR'' after a constitutive promoter (like J23101). According to previous hypothesis, this will provide at steady-state a 'constant concentration' of TetR (we note [TetR]<sub>tot</sub>, and it is supposed to be the TOTAL concentration of TetR, under every form) in the bacterium. If we consider the binding reaction this way (where aTc_-_TetR denotes the complex) |
| - | <center> [[Image: | + | <center> [[Image:aTcTetRn.jpg]]</center> |
| - | with a dissociation constant K, we find at the steady-state | + | with a dissociation constant K<sub>aTc</sub>, we find at the steady-state |
| - | <center> [[Image: | + | <center> [[Image:TetRfree.jpg]]</center> |
| - | where [aTc] denotes the concentration of aTc we introduced in the medium, that will stay constant in all the bacteria along time, assuming that its degradation is near 0, and that the diffusion is quick. | + | where [aTc] denotes the concentration of aTc we introduced in the medium, that will stay constant in all the bacteria along time, assuming that its degradation is near 0, and that the diffusion is quick. |
According to the [[Team:Paris/Modeling#first_Hypothesis|hypothesis '''(3)''']], the activity of ''Ptet'' would verify (keeping the same notations) : | According to the [[Team:Paris/Modeling#first_Hypothesis|hypothesis '''(3)''']], the activity of ''Ptet'' would verify (keeping the same notations) : | ||
| - | <center> [[Image: | + | <center> [[Image:exprTetR.jpg]]</center> |
| - | + | Then, by considering the complexation | |
| - | <center> [[Image: | + | <center> [[Image:TetRPtet.jpg]]</center> |
| - | + | We find at steady-state : | |
| - | + | ||
| - | <center> [[Image: | + | <center> [[Image:HillActPtet.jpg]] </center> |
| - | + | In the last equation, we will have 'access' (see [[Team:Paris/Modeling#first_Hypothesis|hypothesis '''(5)''']] | |
| - | + | ) to [prot]<sub>eq</sub> and possibly to γ<sub>prot</sub>, and we are looking for β<sub>tet</sub>, K<sub>tet</sub> and n<sub>tet</sub>, thanks to our program (see [[Team:Paris/Modeling#getting_a_Hill_function_from_convenient_datas|getting Hill function with convenient datas]]). But we need to know (K<sub>aTc</sub>[TetR<sub>tot</sub>]/(K<sub>aTc</sub> + [aTc]<sup>aTc<sup>)), and we just have [aTc]. However, we could probably get [TetR<sub>tot</sub>], that depends of the constitutive promoter (J23101) we will put before, which we will caracterise (see [[Team:Paris/Modeling/estimation#what_are_we_looking_for_.3F]]). | |
| - | + | Then, we intend to program a new algorithm, based on the same principles of 'findparam' for a classic ''hill function'', but that is looking for more parameters. | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
====<center>By AraC / Arabinose / ''Pbad'' </center>==== | ====<center>By AraC / Arabinose / ''Pbad'' </center>==== | ||
Revision as of 13:43, 18 August 2008
|
 "
"