|
|

| August |
|
|
WET LAB:
Ligation repeat
Performed in order to obtain at least part 1 and part 2 joined
Part 1 |
5 μl |
Part 2 |
5 μl |
Buffer 5x |
4 μl |
T4DNA Pol |
1 μl |
H2O |
5 μl |
Total |
20 μl |
Ligation PCR of P1_P2
H2O |
33 μl |
Buffer |
5 μl |
dNTPs |
2.5 μl |
Oligo 1 up |
2.5 μl |
Oligo 2 low |
2.5 μl |
Mg(Cl)2 |
2.5 μl |
DNA |
1 μl |
Taq |
|
Total |
50 μl |
*The ligation and the PCR were done ___
Plasmid Extraction
Plasmid PBBIMCS_5 was extracted from the cultures of the last day.
PCR ligation Gel of part1_part2

|
WET LAB:
Ligation part1+PRK415
The tube DR of PRK415 was used along with the following samples of part 1:
- DR Part1_9/pJET (130808)
- DR Part1_6/pJET (130808)
- DR Part1_3/pJET (130808)
Recipe for each ligation tube
Vector |
4 μl |
Part 1 |
8 μl |
Buffer 5x |
4 μl |
Enzyme |
1 μl |
H2O |
3 μl |
Restriction
Linearization of pBB to use it as a control in RcnA+pBB Gel
Recipe for each restriction tube:
H20 |
10 μl |
Buffer |
3 μl |
DNA |
16 μl |
EcoR1 |
1 μl |
|
30 μl |
Plasmid Extraction Gel pBB+RcnA
- Molecular Marker (2 μl)
- [1] Cage2 /11_1
- [2] Cage1 /1_2
- [3] Cage2B /1_1
- [4] Cage2a /10_1
- [5] Cage2b /8_1
- [6] Cage2b /6_1
- [7] Cage1 /5_1
- [8] Cage1 /2_1
- [9] Cage2b /4_1

|
1 |
2 |
3 |
4 |
5 |
6 |
Water |
7 μl |
7 μl |
7 μl |
5 μl |
7 μl |
7 μl |
Buffer |
2 μl |
2 μl |
2 μl |
2 μl |
2 μl |
2 μl |
DNA |
10 μl |
10 μl |
10 μl |
12 μl |
10 μl |
10 μl |
Enzyme |
1 μl |
1 μl |
1 μl |
1 μl |
1 μl |
1 μl |
|
20 μl |
20 μl |
20 μl |
20 μl |
20 μl |
20 μl |
*In order to verify the size of the insert in pBB
Extraction plasmid restrictions (pBB+RcnA) with EcoR1
We used:
- [1] Cage2 /11_1
- [2] Cage1 /1_2
- [3] Cage2B /1_1
- [4] Cage2a /10_1
- [5] Cage2b /8_1
- [9] Cage2b /4_1
Gel
- Molecular Marker (2.5 μl)
- Restriction EcoR1 pBBIMCS-5 2b/4 (2 μl)
- Restriction EcoR1 pBBIMCS-5 2a/10 (2 μl)
- Restriction EcoR1 pBBIMCS-5 2b/5 (2 μl)
- Restriction EcoR1 pBBIMCS-5 2a/14(2 μl)
- Restriction EcoR1 pBBIMCS-5 2a/9(2 μl)
- Restriction EcoR1 pBBIMCS-5 limpio con kit(2 μl)
- Restriction EcoR1 BamH1 PRK415 R1(2 μl)
- Restriction EcoR1 BamH1 PRK415 R2(2 μl
- PCR control1 biopart 1 oligo 1up 2low (5 μl)
- PCR control1 biopart 2 oligo 1up 2low (5 μl)
- Negative Control PCR (5 μl)

Conclusions of the previous gel:
- The Oligos 1up and 2low don't join in unexpected sites
- RcnA wasn't inserted in pBB
- It's necessary to repeat the double restriction of PRK
PCR
- Double ligation
- Control without DNA
- Control without oligos (¿Is there something contaminated?)

We run a gel of this three PCRs and we didn't obtain any product.
Repetición PCR
We repeated the previous PCR reaction using the following recipe:
DNA |
2 μl |
H2O |
32 μl |
dNTP’s |
2.5 μl |
Mg(Cl)2 |
2.5 μl |
Oligo 1up |
2.5 μl |
Oligo 2low |
2.5 μl |
Taq |
1 μl |
Buffer |
5 μl |
|
WET LAB
Restrictions gel pBB+RcnA
- Molecular Marker (2.5 μl)
- [1] Cage2 /11_1
- [2] Cage1 /1_2
- [3] Cage2B /1_1
- [4] Cage2a /10_1
- [5] Cage2b /8_1
- [9] Cage2b /4_
- pBB1
- pBB2
- pBB3

Cultured from a strain result of the transformation
|
We received an e-mail from the Mexico-UNAM-IPN with several questions in regards to our project, here is our reply:
Hello,
We apologize for the late reply, but we had to discuss carefully our answer.
First of all, we think you are confused about what our project really is. We want to make bacteria to modify the extracellular nickel concentration in response to an external signal (AHL in this case), and of course, be able to predict to what extent the concentration of the input signal will affect the amount of nickel in the medium. To achieve this, it is true we have to synchronize our cell population at the beginning. This is easy to do and doesn't represent any technical problems.
We are very conscious of the facts you tell us, first: we know the half-life of the lactones is relatively long (24 hrs as you say). That's why we are including AiiA under a constitutive promoter in our model, which degrades AHL very efficiently. This will ensure AHL does not saturate the medium. Second, we know AiiA does not diffuse freely through the cell membrane. However, we don't need that to happen, as each cell will degrade its own AHL (yes, we are assuming that all AHL will enter a cell within a window of time).
In other words, we do not need to synchronize the bacterial population more than in the first step. We are considering that some cells may respond earlier than others. However, we are assuming that, as we are not changing the physical nor chemical conditions, the proportion of cells responding "earlier" will remain constant, thus allowing us to draw some conclusions of the behaviour of the population as a whole. We hope you see why the synchrony is no longer important for our project.
To summarize what we plan to do, AHL will enter the cell and form a dimer with LuxR (which is under a constitutive promoter, so AHL is the only limiting step). This will start the transcription of cI*, which will repress the expression of RcnA. RcnA is the nickel efflux pump, and thus we are aiming to predict the amount of AHL necessary to get the desired extracellular nickel concentration.
We are doing small moves. At first, we only want to make one successful assay. We hope that in the near future we will be able to use the response time of the system to generate a succession of desired nickel concentrations, thus generating a song.
We hope this letter answers your questions,
LCG-UNAM-Mexico Team
Cuernavaca, Morelos
MODELING:
Reaction 3, AHL:LuxR
Conflict: k3 (ON) <k3 (OFF)?
Reference: Goryachev et al. (2006)
The references they cite for obtaining their parameters were not specific for that kind of parameters, in fact, one mentions the rate of RNA polymerase in HUMAN!
- Check whether the article mentions how they transformed the parameters.
They seem to explain why in their model, the k3(ON) is "so small" to start with:
<<common to all models considered here, is that the stability of the state "off" defined by the constitutive Transcription levels of I and R comes at a price of high value for the critical self Extracellular concentration.>>
This explains the criteria they used to determine their parameters:
<<For each layout we attempted to identify a set of parameters that optimize the functional fitness of the network. The search in the parameter space is constrained by requesting that the kinetic parameters must remain in the biologically realistic range and the resulting network should demonstrate the behavior compatible with our present understanding of the phenomenon quorum sensing.>>
WET LAB
Agarose Gel 1% low fusion point for band purification
- Gel1
- Molecular Weight marker
- [1]
- [2]
- [3]
Ligation PCR parte1+parte2(35 ciclos)
- 1_2 oligo 1up-2lower
- 1_1 oligo 1up-1lower
- 2_2 oligo 2up-2lower
- 1_3 oligo 1up-3lower
- 1_2 oligo 1up-2lower
Recipes:
DNA |
2 μl |
Water |
32 μl |
dNTP’s |
2.5 μl |
Mg(Cl)2 |
2.5 μl |
Oligo 1up |
2.5 μl |
Oligo 2low |
2.5 μl |
Taq |
1 μl |
Buffer |
5 μl |
|
MODELING:
Reaction 4: Natural degradation of cI*
Half life of cI*:
The half-life of a modified cI is of 4 minutes, according to Elowitz & Leibler (2000). They analyze the LAA tail, and JB Andersen et al (1998) conclude that the LAA and LVA tails confer about the same time of life to GFP.
Reaction rate:
Once we get the half-life time of the protein, how do we calculate the rate of reaction and the flow?
The half-life of a reaction (t1/2) is the time it takes for half of the reagents to become products. In a first order reaction, t1/2 is a constant and can be calculated from the rate constant, as follows:
t1/2 =-ln(0.5)/k=0.693/k
This reciprocal relationship between the half life time and the rate constant is very useful when making an estimate of the time a reaction will take to occur. Thus, for k = 0.01/s, the half life time would be about 70 s. For k = 10/s, the half life would be of about 0.07 s or 70 milliseconds. The average life of the reactions of first order is also independent of the initial concentration. If the first half of the molecules react in aprox 20 s, half of the remaining molecules will also take 20 s to react, and so on. The fact that the average lifetime in an unimolecular reaction is a constant means that, at any time of the reaction, a constant fraction of reactive molecules have enough energy to overcome the kinetic barrier and become molecules of product. This makes sense because the energy of a set of molecules is distributed randomly according to a Boltzmann distribution.
* With information from RT Sauer (1999).
NOTE: A first order reaction is the type A → B.
Today we sent an e-mail to Dr. Peter Chivers, an expert in RcnR, to ask him for information in regards to RcnA.
Our mail to Dr. Peter Chivers:
Dr. Peter Chivers,
Hi! My name is Carlos and I'm a student of the Undergraduate Program in Genomic Sciences of the National Autonomous University of Mexico and currently I'm on the fourth year.
The main reason of this e-mail is to get in contact with you, I'll explain you why as briefly as I can. Several students of the program decided to participate in a systems biology competition known as iGEM, and our project focuses on regulating nickel efflux in e. coli. We have to both build the circuit and design a mathematical model to accurately predict the behaviour of our system.
Taking several issues in consideration we decided to regulate RcnA.
Here is the page of our team in case you're interested in reading more about our project:
https://2008.igem.org/Team:LCG-UNAM-Mexico
However we've been having some trouble in finding parameters for our model (such as RcnA's half-life, efficiency, synthesis rate among others). Therefore, having exhausted (or so we believe) the literary resources and having found your name in many of the papers regarding RcnA we decided to contact you. I'll try to explain with more detail the parameters we still lack in case you're interested in providing us with information.
I greatly thank you for taking the time to read this e-mail and await your answer.
Carlos Vargas
Dr. Peter Chivers' reply
Dear Carlos,
Thank you for your e-mail. From what I read on your website, it looks like your team is tackling an interesting project. Unfortunately, there are very few papers on RcnA itself, and none of them have examined the parameters that you need for your model. As far as I know, the groups that have published in the RcnR/RcnA area don't typically do those sorts of measurements so they are unlikely that have the data you need. We have no plans to do these experiments as my lab focuses on the nickel responsive regulators, RcnR and NikR.
I'm sorry that I can't be of more assistance. Good luck with your project, I hope it is well received at the iGEM competition.
Best regards,
Peter
WET LAB
Agarose Gel 1% low fusion point for band purification
- Molecular Weight Marker
- [1] Cage2 /11_1 pBB+RcnA
- [2] Cage1 /1_2 pBB+RcnA
- [5] Cage2b /8_1 pBB+RcnA
- [9] Cage2b /4_1 pBB+RcnA
Gel Purification PCR of pBB+rcnA
- Purification 1
- Purification 2
- Purification 3
- Purification 4 clean PRK
- Positive control
- Negative control
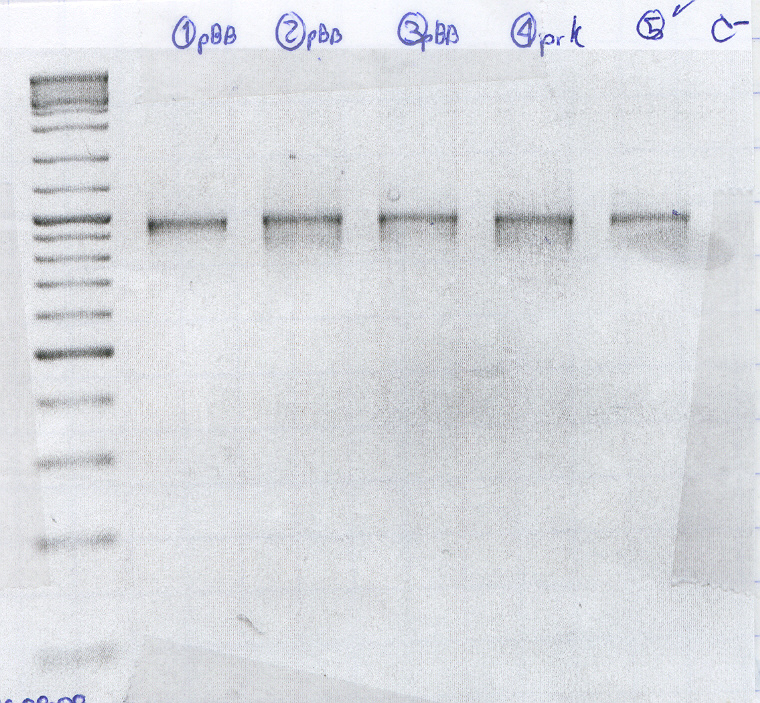
The cells seem to carry the plasmid with RcnA, However one of the negative controls gave product.
All of them were positive, however there are some spurious products.(no photo was added because the low melting gels are quite fragile and we couldn't carry it to the transiluminator with camera.)
Recipe:
DNA |
2 μl |
Water |
32 μl |
dNTP’s |
2.5 μl |
Mg(Cl)2 |
2.5 μl |
Oligo 1up |
2.5 μl |
Oligo 2low |
2.5 μl |
Taq |
1 μl |
Buffer |
5 μl |
A gel with the extraction of the plasmid PRK415+part_1 was run.
Gel Electrophoresis of PCR_Ligation part1+part2
- [5] 1_2 oligo 1up-2lower
- [4] 1_2 oligo 1up-2lower
- [3] 1_2 oligo 1up-2lower
- Molecular marker
- [2] 1_2 oligo 1up-2lower
- [1] 1_2 oligo 1up-2lower

We have got part1+part2 ligation
|
WET LAB
PRK+part1 PCR
Recipe:
DNA |
1 μl |
11 μl |
H2O |
33 μl |
363 μl |
dNTP’s |
2.5 μl |
27.5 μl |
Mg(Cl)2 |
2.5 μl |
27.5 μl |
Oligo 1up |
2.5 μl |
27.5 μl |
Oligo 2low |
2.5 μl |
27.5 μl |
taq |
1 μl |
11 μl |
Buffer |
5 μl |
55 μl |
|
50 μl |
550 μl |
PCR gel of RcnA (Obtained of the purification in gel of pBB + RcnA)
1.-Molecular-weight marker
2.-[1] RcnA
3.-[2] RcnA
4.-[5] RcnA
5.-[9] RcnA
6.-Control +
7.-Control -
Ligation part1+part2 (rttH) PCR
Reaction 1 |
|
DNA |
2 μl |
H2O |
10 μl |
dNTP’s |
4 μl |
Mg(Ac)2 |
3 μl |
Oligo 1up |
2.5 μl |
Oligo 2low |
2.5 μl |
Buffer |
6 μl |
Reaction 2 |
|
H2O |
9 μl |
Buffer |
9 μl |
RttH |
11 μl |
Ligation part1_part2 Gel PCR
1.-Molecular-weight marker
2.-PCR Lp1+P2 1.1
3.- PCR Lp1+P2 1.2
4.- PCR Lp1+P2 5.1
5.- PCR Lp1+P2 5.2
6.-Negative control
7.-PRK + part 1_1
8.- PRK + part1_2
PCR gel of RcnA (Obtained of the purification in gel of pBB + RcnA) Repetition
1.- Molecular-weight marker
2.-[1] RcnA
3.-[2] RcnA
4.-[5] RcnA
5.-[9] RcnA
6.-Control +
7.-Control –
Ligation part1+part2 Restriction with BamH1
|
For1_1 |
For1_2 |
H2O |
10 μl |
5 μl |
Buffer2 |
2 μl |
2 μl |
BSA |
2 μl |
2 μl |
DNA |
5 μl |
10 μl |
Enzime(BamH1) |
1 μl |
1 μl |
|
20 μl |
20 μl |
PRK+part1 Gel PCR
1.- Molecular-weight marker
2.-[3]
3.-[4]
4.-[5]
5.-[6]
6.-[7]
7.-[9]
8.-[10]
9.-c-
PCRs
With oligos 4 up, 4 lower
1.- pBB+RcnA 1 band purification
2.- pBB+RcnA 1 band purification
5.- pBB+RcnA 1 band purification
9.- pBB+RcnA 1 band purification
-1.-Negative control 1(prk)
-2.-Negative control 2(pBB)
+ Positive control (PCR August 22)
With oligos 1 up, 3 lower
L1 Ligation part1+part2 1_2
L- Negative control
DNA |
1 μl |
H2O |
33 μl |
dNTP’s |
2.5 μl |
Mg(Cl)2 |
2.5 μl |
Oligo 1up |
2.5 μl |
Oligo 2low |
2.5 μl |
Taq |
1 μl |
Buffer |
5 μl |
|
WET LAB
2% Agarose gel with the next samples:
1.- Molecular-weight marker
2.-[1] pBB+RcnA 1 band purification
3.-[2] pBB+RcnA 1 band purification
4.-[5] pBB+RcnA 1 band purification
5.-[9] pBB+RcnA 1 band purification
6.-[-1]Negative control 1(prk)
7.-[-2]Negative control 2(pBB)
8.-[+ ]Positive control (PCR August 22)
9.- L1 Ligation part1+part2 1_2
10.- - Negative control
11.-1_1 Restriction BamH 220808 part1+part2
12.-1_2 Restriction BamH 220808 part1+part2
13.- Molecular marker
* 5 μl of each sample.

Ligation part1+part2 restrictions
H2O |
4 μl |
Buffer EcoR1 |
5 μl |
BSA |
5 μl |
DNA |
30 μl |
Xba1 enzime |
3 μl |
EcoR1 enzime |
3 μl |
PRK415 Restriction
H2O |
5 μl |
Buffer EcoR1 |
3 μl |
BSA |
3 μl |
DNA |
15 μl |
Xba1 Enzime |
2 μl |
EcoR1 Enzime |
2 μl |
(part1+part2)+PRK415 Ligation
A quick ligation kit was used.
Vector (PRK415) |
15 μl |
Part1+Part2 |
7 μl |
Buffer 5X |
8 μl |
Enzime |
2 μl |
H2O |
8 μl |
|
40 μl |
(part1+part2)+ part3 Ligation
A quick ligation kit was used.
Part3 |
10 μl |
Part1+Part2 |
10 μl |
Buffer 5X |
8 μl |
Enzime |
2 μl |
H2O |
10 μl |
|
40 μl |
|
MODELING:
Checking parameters:
Reaction 6
The value that had been found before for k6 (Pl) = 0.20mM / h is in reality the flow of the reaction (which is why the units are mM/h). That is, how many mRNA molecules are being produced per unit of time. The system in which they measured this parameter is a derivative of the plasmid pBR322, pTrc99A. It has the same origin of replication and the number of copies is estimated at 15-20 but under certain conditions where replication is limited it appears to be between 3-5 copies.
Using the previous information we can calculate the value of k+ of the reaction in our system if we consider that each promoter acts independently and we multiply by the ratio between the number of copies of our plasmid and those of their plasmid.
WET LAB
PCR, ligation (1_2)_3
1.-PCR (1_2)_3 oligo 1up 3 low
2.-PCR (1_2)_3M oligo 1up 3 low
3.-c- negative control (no DNA)
4.- pcr (1_2)_3N oligo 2up 3 low control
*Independently added to each tube.
*The alignment temperature was raised 5ºC.
*Problem in oligo 3 low.
|
One reaction |
Four |
DNA |
1 μl |
4μl |
H2O |
33 μl |
132μl |
dNTP’s |
2.5 μl |
10μl |
Mg(Cl)2 |
2.5 μl |
10μl |
Oligo 1up |
2.5 μl |
10μl |
Oligo 2low |
2.5 μl |
10μl |
taq |
1 μl |
4 μl |
Buffer |
5 μl |
20 μl |
|
50 μl |
200μl |
|
WET LAB
1% Agarose Gel
1.- Molecular-weight marker
2.-[1] part (1_2)_3 oligo 1up_3low
3.-[2] part(1_2)_3M oligo 1up 3low
4.-[3] Negative control (no DNA)
5.-[4] Negative control part(1_2)_3N
Vector PRK415 restriction to ligate (p1+p2)
|
Simple restriction Pst1 |
Double restriction EcoR1/Pst1 |
H2O |
3 μl |
6 μl |
Buffer 10X |
4 μl |
2 μl |
DNA |
30 μl |
10 μl |
Pst1 |
3 μl |
1 μl |
EcoR1 |
0 μl |
1 μl |
|
40 μl |
20 μl |
Ligation [p1_p2]-p3 restriction
Final volume 70 μl
H2O |
9 μl |
Buffer 10X |
7 μl |
DNA |
50 μl |
Pst1 |
2 μl |
EcoR1 |
2 μl |
|
70 μl |
Electrophoresis, double restriction, band purification pBB+RcnA
1.- Molecular-weight marker
2.-[1]
3.-[2]
4.-[5]
5.-[9]

|
MODELING:
To-do List
1. Find Missing Parameters.
- cI Dimerization.
- Nickel efflux . (Estimate?).
- RcnA degradation.
- Nickel Internalization (Experimentally,?).
- Initial concentrations (aiiA, LuxR? (Define arbitrary medium), ? (by number of copies of the plasmid; change to molar), Ni-ext (experimental)).
2. Review tools in SimBiology (sensitivity analysis, parameter estimation, moiety conservation).
3. Stationary states of the system (whether there is multistationarity).
4. Jacobian.
5. Analysis of the stechiometric matrix (also analyze the null space and its transpose).
6. Electrochemical theory (the relationship between electric potential and the concentration of nickel).
7. Electrodes.
->We need to check for advances in the construction of the measurement device we will use.
->Take into account the possibility of asking for support to Dr. Peña.
WET LAB
Streak
Strains streak of PRK415+(part1_part2) in tube for plasmid extraction (verification)
Transformation
Transformation on ligartions part1_pat2_part3 in DH5α
Restrictions
|
Bioparts |
Plasmid |
H2O |
11μl |
6 μl |
Buffer 10X |
5 μl |
2 μl |
DNA |
30 μl |
20 μl |
Pst1 |
2 μl |
1 μl |
EcoR1 |
2 μl |
1 μl |
|
50μl |
30 μl |
Ligation
Two ligations of every version of the ligation part1+part2+part3 were made. 25 μl of insert and 25 μl of plasmid were added to each one.
DNA insert |
25 μl |
PRK415 |
25 μl |
Buffer T4 ligase 10X |
6 μl |
H2O |
3 μl |
T4 ligase |
1 μl |
|
60 μl |
|
WET LAB
Plasmid ligation p1+p2+PRK415 gel.
No results.
TO DO's:charge and run a gel
|
|


|
| |



