Team:UNIPV-Pavia/Notebook/Week6
From 2008.igem.org
Notebook
| Week 1 | Week 2 | Week 3 | Week 4 | Week 5 | Week 6 | Week 7 |
|---|---|---|---|---|---|---|
| Week 8 | Week 9 | Week 10 | Week 11 | Week 12 | Week 13 | Week 14 |
| Week 15 | Week 16 | Week 17 | Week 18 | Week 19 | Week 20 | Week 21 |
| Week 22 | Week 23 | Week 24 |
Week 6: 06/23/08 - 06/27/08
06/23/08
- Colony PCR for BBa_B0030-BBa_C0061 and BBa_B0030 (no Ant.Phosph.)-BBa_C0061: 10 colonies for every plate.
- We ran PCR results: some colonies showed both ligated plasmid (heavier band) and false positive plasmid, because it was difficult to pick up single colonies from bacteria carpet. Ligation with Antarctic Phosphatase showed non-pure colonies and 3 false positives. Ligation without Antarctic Phosphatase showed 3 non-pure colonies and 7 pure colonies.
- We decided to avoid Antarctic Phosphatase treatment in next ligations to save time.
- We also decided to cut up to 1 µg of vector, to reduce background noise.
- We choose 4th and 7th colonies to grow 9 ml cultures overnight.
- We also infected 9 ml LB + Amp with 30 µl of:
| BBa_E0240 | BBa_C0078 | BBa_B0030 | BBa_J23100 |
- glycerol stocks. We incubated all the 6 cultures at 37°C, 220 rpm.
- We transformed 10 µl of the remaining ligations:
- BBa_B0030-BBa_E0040
- BBa_B0030-BBa_E1010
- BBa_B0030-BBa_C0051
- Wiki updating: Project section.
06/24/08
- All the 3 ligation plates showed carpets. We decided to streak plates to produce single colonies plates. We incubated the 3 single colonies plates at 37°C overnight.
- Glycerol stocks for:
| BBa_E0240 | BBa_C0078 | BBa_B0030-BBa_C0061(4) |
| BBa_J23100 | BBa_B0030 | BBa_B0030-BBa_C0061(7) |
- Miniprep for these 6 plasmids.
- Plasmid digestion:
| BBa_E0240 (X-P) | BBa_C0078 (X-P) |
| BBa_J23100 (S-P) | BBa_B0030 (S-P) |
- DNA precipitation with sodium acetate for BBa_J23100 (S-P) and BBa_B0030 (S-P).
- Gel run for BBa_E0240 (X-P) and BBa_C0078 (X-P)
- Gel extraction.
- Ligation:
- BBa_J23100-BBa_E0240
- BBa_B0030-BBa_C0078
06/25/08
- We transformed 5 µl of the two ligations.
- We also transformed 0.5 µl of BBa_B0030(S-P) and BBa_J23100(S-P) plasmids to estimate background noise.
- Colony PCR (6 colonies for each plate) for:
- BBa_B0030-BBa_E0040
- BBa_B0030-BBa_E1010
- BBa_B0030-BBa_C0051
- The gel showed many working ligations! we choose the 1st colony for BBa_B0030-BBa_E0040, the 6th colony for BBa_B0030-BBa_C0051 and the 2nd colony for BBa_B0030-BBa_E1010 to grow 9 ml LB + Amp overnight cultures.
06/26/08
- Glycerol stocks for:
- BBa_B0030-BBa_E0040 (1)
- BBa_B0030-BBa_E1010 (6)
- BBa_B0030-BBa_C0051 (2)
- Miniprep for these 3 ligations. We sent purified plasmids to Primm for sequencing.
- BBa_J23100-BBa_E0240 and BBa_B0030-BBa_C0078 plates showed a carpet.
- We prepared single colonies plates for BBa_J23100-BBa_E0240 and BBa_B0030-BBa_C0078.
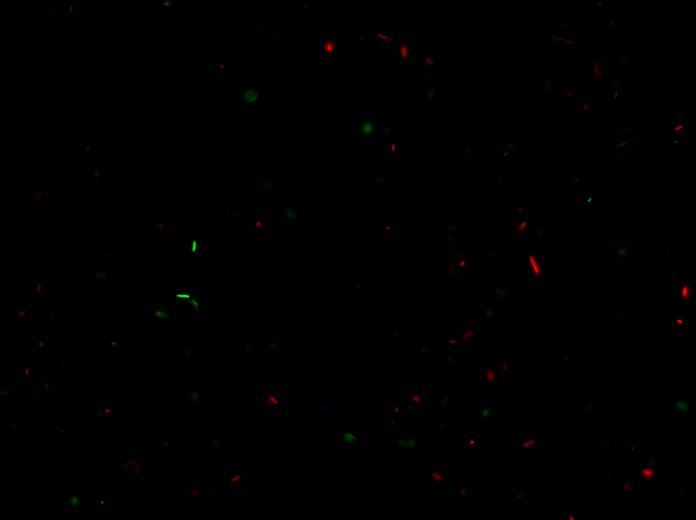
- NOTE: BBa_J23100-BBa_E0240 plate showed red colonies (false positives) and normal color colonies (ligations).
- We tested fluorescence for BBa_J23100-BBa_E0240: we streaked the plate and infected 100 µl of LB + Amp. We incubated this culture for 2 hours at 37°C, 220 rpm. Then we watched green, blue and red fluorescence channels at microscope. Some cells expressed GFP (cells with ligated plasmid) and some cells expressed RFP (false positives).
06/27/08
We put BBa_J23100-BBa_E0240 and BBa_B0030-BBa_C0078 single colonies plates at +4°C. Next week we will perform colony PCR for these two ligations.
 "
"