Team:UNIPV-Pavia/Notebook/Week8
From 2008.igem.org
(Difference between revisions)
| (22 intermediate revisions not shown) | |||
| Line 38: | Line 38: | ||
!|[[Team:UNIPV-Pavia/Notebook/Week13|Week 13]] | !|[[Team:UNIPV-Pavia/Notebook/Week13|Week 13]] | ||
!|[[Team:UNIPV-Pavia/Notebook/Week14|Week 14]] | !|[[Team:UNIPV-Pavia/Notebook/Week14|Week 14]] | ||
| + | |- | ||
| + | !|[[Team:UNIPV-Pavia/Notebook/Week15|Week 15]] | ||
| + | !|[[Team:UNIPV-Pavia/Notebook/Week16|Week 16]] | ||
| + | !|[[Team:UNIPV-Pavia/Notebook/Week17|Week 17]] | ||
| + | !|[[Team:UNIPV-Pavia/Notebook/Week18|Week 18]] | ||
| + | !|[[Team:UNIPV-Pavia/Notebook/Week19|Week 19]] | ||
| + | !|[[Team:UNIPV-Pavia/Notebook/Week20|Week 20]] | ||
| + | !|[[Team:UNIPV-Pavia/Notebook/Week21|Week 21]] | ||
| + | |- | ||
| + | !|[[Team:UNIPV-Pavia/Notebook/Week22|Week 22]] | ||
| + | !|[[Team:UNIPV-Pavia/Notebook/Week23|Week 23]] | ||
| + | !|[[Team:UNIPV-Pavia/Notebook/Week24|Week 24]] | ||
|} | |} | ||
| Line 95: | Line 107: | ||
|BBa_I15010 | |BBa_I15010 | ||
|BBa_B0030-BBa_E0040-'''BBa_B1006''' (1) | |BBa_B0030-BBa_E0040-'''BBa_B1006''' (1) | ||
| - | |||
|- | |- | ||
|BBa_C0012 | |BBa_C0012 | ||
|BBa_R0062 | |BBa_R0062 | ||
|BBa_B0030-BBa_C0051-'''BBa_B0030''' | |BBa_B0030-BBa_C0051-'''BBa_B0030''' | ||
| + | |- | ||
|BBa_J23100-'''BBa_B0030''' | |BBa_J23100-'''BBa_B0030''' | ||
| + | |BBa_B0030-BBa_E1010-'''BBa_B1006''' (1) | ||
|} | |} | ||
| Line 108: | Line 121: | ||
{|cellpadding="20px" | {|cellpadding="20px" | ||
|BBa_B0030 (S-P) | |BBa_B0030 (S-P) | ||
| - | |||
|BBa_B0030-BBa_E0040-'''BBa_B1006''' (1) (E-X) | |BBa_B0030-BBa_E0040-'''BBa_B1006''' (1) (E-X) | ||
|BBa_B0030-BBa_E1010-'''BBa_B1006''' (1) (E-X) | |BBa_B0030-BBa_E1010-'''BBa_B1006''' (1) (E-X) | ||
|- | |- | ||
|BBa_C0012 (X-P) | |BBa_C0012 (X-P) | ||
| - | |||
|BBa_B0030-BBa_C0051-'''BBa_B0030''' (S-P) | |BBa_B0030-BBa_C0051-'''BBa_B0030''' (S-P) | ||
|BBa_J23100-'''BBa_B0030''' (E-S) | |BBa_J23100-'''BBa_B0030''' (E-S) | ||
|- | |- | ||
| + | |BBa_I15010 (X-P) | ||
|BBa_J23100-'''BBa_B0030'''-BBa_C0040 (E-S) | |BBa_J23100-'''BBa_B0030'''-BBa_C0040 (E-S) | ||
|'''BBa_R0051'''-BBa_B0030-BBa_C0062 (E-S) | |'''BBa_R0051'''-BBa_B0030-BBa_C0062 (E-S) | ||
| + | |- | ||
| + | |BBa_R0062 (E-X) | ||
|BBa_B0030-BBa_C0061-'''BBa_B1006''' (E-S) | |BBa_B0030-BBa_C0061-'''BBa_B1006''' (E-S) | ||
|} | |} | ||
| - | *Gel run/cut | + | *Gel run/cut/gel extraction for: |
| + | {|cellpadding="20px" | ||
| + | |BBa_I15010 (X-P) | ||
| + | |BBa_J23100-'''BBa_B0030''' (E-S) | ||
| + | |BBa_J23100-'''BBa_B0030'''-BBa_C0040 (E-S) | ||
| + | |- | ||
| + | |BBa_C0012 (X-P) | ||
| + | |'''BBa_R0051'''-BBa_B0030-BBa_C0062 (E-S) | ||
| + | |BBa_B0030-BBa_C0061-'''BBa_B1006''' (E-S) | ||
| + | |} | ||
| - | * | + | *DNA precipitation with sodium acetate for: |
| + | {|cellpadding="20px" | ||
| + | |BBa_B0030 (S-P) | ||
| + | |BBa_B0030-BBa_E0040-'''BBa_B1006''' (1) (E-X) | ||
| + | |BBa_B0030-BBa_E1010-'''BBa_B1006''' (1) (E-X) | ||
| + | |- | ||
| + | |BBa_R0062 (E-X) | ||
| + | |BBa_B0030-BBa_C0051-'''BBa_B0030''' (S-P) | ||
| + | |} | ||
*(We already had BBa_I15009(X-P) and BBa_B1006(E-X)) | *(We already had BBa_I15009(X-P) and BBa_B1006(E-X)) | ||
| Line 145: | Line 176: | ||
<br><br> | <br><br> | ||
'''07/9/08''' | '''07/9/08''' | ||
| + | <br> | ||
| + | *We transformed the 9 ligations (2 µl) and plated transformed bacteria. We incubated plates at 37°C ovenight. | ||
| + | |||
| + | *Glycerol stocks for BBa_B1006 and '''BBa_B0030'''-BBa_C0078. | ||
| + | |||
| + | *Miniprep for BBa_B1006 and '''BBa_B0030'''-BBa_C0078. | ||
| + | |||
| + | *Plasmid digestion: | ||
| + | {|cellpadding="20px" | ||
| + | |BBa_B1006 (E-X) | ||
| + | |'''BBa_B0030'''-BBa_C0078 (E-S) | ||
| + | |} | ||
| + | |||
| + | *Gel run/cut/gel extraction for '''BBa_B0030'''-BBa_C0078 (E-S). | ||
| + | |||
| + | *DNA precipitation with sodium acetate for BBa_B1006 (E-X). | ||
| + | |||
| + | *Ligation: BBa_B0030-BBa_C0078-'''BBa_B1006''' | ||
| + | |||
| + | *We incubated ligation at 16°C overnight. | ||
| + | |||
| + | <br><br> | ||
| + | '''07/10/08''' | ||
| + | <br> | ||
| + | *We transformed BBa_B0030-BBa_C0078-'''BBa_B1006''' ligation (2 µl) and plated transformed bacteria. We incubated the plate at 37°C ovenight. | ||
| + | |||
| + | *All the ligation plates showed colonies! There were carpets, but we could pick up some single colonies for colony PCR. | ||
| + | |||
| + | *Colony PCR for: | ||
| + | #'''BBa_B0030'''-BBa_I15009 | ||
| + | #BBa_J23100-BBa_B0030-BBa_C0040-'''BBa_B1006''' | ||
| + | #BBa_R0051-BBa_B0030-BBa_C0062-'''BBa_B1006''' | ||
| + | #BBa_B0030-BBa_C0051-'''BBa_B0030'''-BBa_C0079 | ||
| + | #BBa_B0030-BBa_C0061-BBa_B1006-'''BBa_R0062''' | ||
| + | #BBa_J23100-'''BBa_B0030'''-BBa_C0012 | ||
| + | #BBa_J23100-'''BBa_B0030'''-BBa_I15010 | ||
| + | |||
| + | {| | ||
| + | |[[Image:pv_falcon_colonypcr_9_7_08.jpg|thumb|300px|left|1 ml of infected LB + antibiotic for all the colonies we picked up to perform colony PCR]] | ||
| + | ||[[Image:pv_pcr_9_7_08.jpg|thumb|300px|left|Colony PCR]] | ||
| + | |} | ||
| + | |||
| + | {| | ||
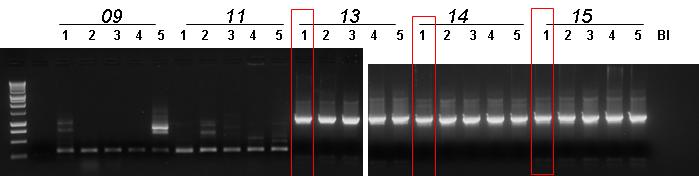
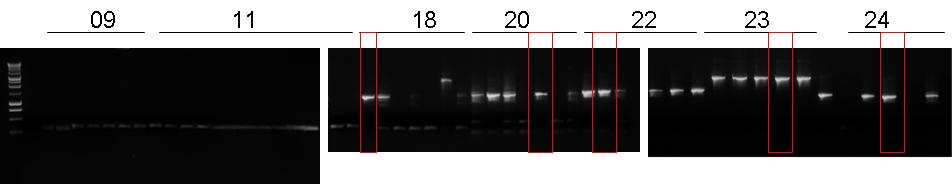
| + | |[[Image:pv_pcr_9-11-18-20-22-23-24.jpg|thumb|850px|left|Colony PCR: Marker (1), BBa_J23100-'''BBa_B0030'''-BBa_C0012, BBa_J23100-'''BBa_B0030'''-BBa_I15010, '''BBa_B0030'''-BBa_I15009, BBa_J23100-BBa_B0030-BBa_C0040-'''BBa_B1006''', BBa_R0051-BBa_B0030-BBa_C0062-'''BBa_B1006''', BBa_B0030-BBa_C0051-'''BBa_B0030'''-BBa_C0079, BBa_B0030-BBa_C0061-BBa_B1006-'''BBa_R0062''']] | ||
| + | |} | ||
| + | |||
| + | *Gel results: | ||
| + | #'''BBa_B0030'''-BBa_I15009 1st colony was chosen. | ||
| + | #BBa_J23100-BBa_B0030-BBa_C0040-'''BBa_B1006''' 5th colony was chosen. | ||
| + | #BBa_R0051-BBa_B0030-BBa_C0062-'''BBa_B1006''' 2nd colony was chosen. | ||
| + | #BBa_B0030-BBa_C0051-'''BBa_B0030'''-BBa_C0079 4th colony was chosen. | ||
| + | #BBa_B0030-BBa_C0061-BBa_B1006-'''BBa_R0062''' 3rd colont was chosen. | ||
| + | #BBa_J23100-'''BBa_B0030'''-BBa_C0012 did not show true positive colonies. | ||
| + | #BBa_J23100-'''BBa_B0030'''-BBa_I15010 did not show true positive colonies. | ||
| + | |||
| + | *We streaked BBa_J23100-BBa_B0030-BBa_E0040-'''BBa_B1006''' and BBa_J23100-BBa_B0030-BBa_E1010-'''BBa_B1006''' with a top to test the parts (NOTE: we could see some red colonies on BBa_J23100-BBa_B0030-BBa_E1010-'''BBa_B1006''' plate!these colonies correspond to bacteria with correctly ligated plasmid, while normal color colonies correspond to bacteria with BBa_B0030-BBa_E1010-'''BBa_B1006''' plasmid, without constitutive promoter). | ||
| + | **We infected 100 µl LB + Amp with the tips and incubated the culture at 37°C, 220 rpm for 2 hours. | ||
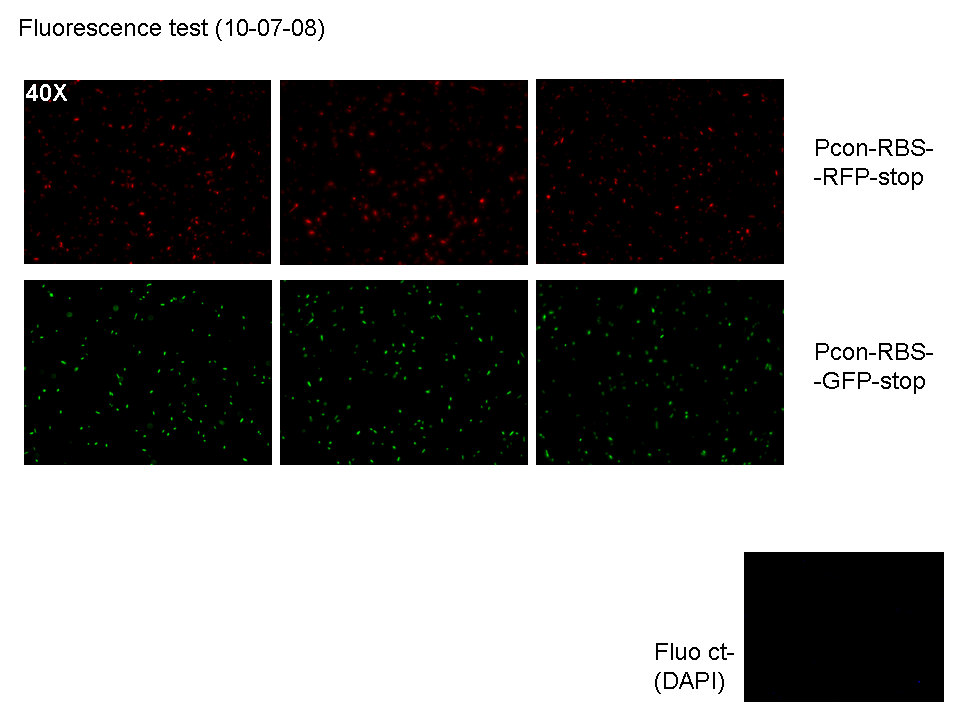
| + | **Then we watched green, blue and red fluorescence channels at microscope (50 µl of each culture): | ||
| + | |||
| + | {| | ||
| + | |[[Image:pv_G_R_10_7_08.png|thumb|700px|left|3 acquisitions for red cells; 3 acquisition for green cells; one acquisition for DAPI channel (blue) for green cells to check for impurities (DAPI channel acquisition for red cells was not saved, but it didn't show any blue area)]] | ||
| + | |} | ||
| + | |||
| + | *LB preparation: 0.5 l LB + Amp for plates. | ||
| + | |||
| + | {| | ||
| + | |[[Image:pv_LB_tower_10_7_08.jpg|thumb|300px|left|Prepared LB plates tower]] | ||
| + | |} | ||
| + | |||
| + | *BBa_R0010 resuspension from Registry of Standard Parts. | ||
| + | |||
| + | *We transformed (4 µl) BBa_R0010 and plated transformed bacteria. We incubated BBa_R0010 plate at 37°C overnight. | ||
| + | |||
| + | *We infected 9 ml LB + suitable antibiotic with 30 µl of these glycerol stocks: | ||
| + | {|cellpadding="20px" | ||
| + | |BBa_C0012 | ||
| + | |'''BBa_B0030'''-BBa_I15009 (1) | ||
| + | |- | ||
| + | |BBa_J23100-BBa_B0030-BBa_C0040-'''BBa_B1006''' (5) | ||
| + | |BBa_R0051-BBa_B0030-BBa_C0062-'''BBa_B1006''' (2) | ||
| + | |- | ||
| + | |BBa_J23100-'''BBa_B0030''' | ||
| + | |BBa_B0030-BBa_C0051-'''BBa_B0030'''-BBa_C0079 (4) | ||
| + | |- | ||
| + | |BBa_B0030-BBa_C0061-BBa_B1006-'''BBa_R0062''' (3) | ||
| + | |} | ||
| + | |||
| + | *We also picked up one colony from BBa_I15010 plate with a tip and infected 9 ml LB + Kan. We incubated the 8 cultures at 37°C, 220 rpm overnight. | ||
| + | |||
| + | <br><br> | ||
| + | '''07/11/08''' | ||
| + | <br> | ||
| + | *BBa_R0010 plate showed 2 colonies. We picked up one colony and infected 9 ml LB + Amp to grow an overnight culture. | ||
| + | |||
| + | *BBa_B0030-BBa_C0078-'''BBa_B1006''' plate showed many colonies. We used 7 colonies to perform colony PCR. | ||
| + | |||
| + | {| | ||
| + | |[[Image:pv_pcr_17.png|thumb|600px|left|7 colonies for BBa_B0030-BBa_C0078-'''BBa_B1006''']] | ||
| + | |} | ||
| + | |||
| + | *Gel results: all the 7 colonies contained ligated plasmid; we chose the 5th colony to grow a 9 ml overnigth culture because 5th it showed no impurities. | ||
| + | |||
| + | *Glycerol stocks for: | ||
| + | {|cellpadding="20px" | ||
| + | |BBa_C0012 | ||
| + | |'''BBa_B0030'''-BBa_I15009 (1) | ||
| + | |- | ||
| + | |BBa_I15010 | ||
| + | |BBa_J23100-BBa_B0030-BBa_C0040-'''BBa_B1006''' (5) | ||
| + | |- | ||
| + | |BBa_J23100-'''BBa_B0030''' | ||
| + | |BBa_R0051-BBa_B0030-BBa_C0062-'''BBa_B1006''' (2) | ||
| + | |- | ||
| + | |BBa_B0030-BBa_C0051-'''BBa_B0030'''-BBa_C0079 (4) | ||
| + | |BBa_B0030-BBa_C0061-BBa_B1006-'''BBa_R0062''' (3) | ||
| + | |} | ||
| + | |||
| + | *Miniprep for these 8 parts. | ||
| + | |||
| + | *We sent BBa_I15010 and BBa_B0030-BBa_C0061-BBa_B1006-'''BBa_R0062''' (3) to Primm for sequencing. | ||
| + | |||
| + | *Plasmid digestion for: | ||
| + | {|cellpadding="20px" | ||
| + | |BBa_C0012 (X-P) | ||
| + | |'''BBa_B0030'''-BBa_I15009 (1) (E-S) | ||
| + | |BBa_B1006 (we already had this plasmid at -20°C) (E-P) | ||
| + | |- | ||
| + | |BBa_J23100-'''BBa_B0030''' (E-S) | ||
| + | |BBa_J23100-BBa_B0030-BBa_C0040-'''BBa_B1006''' (5) (E-S) | ||
| + | |BBa_R0051-BBa_B0030-BBa_C0062-'''BBa_B1006''' (2) (E-S) | ||
| + | |- | ||
| + | |BBa_J23100-'''BBa_B0030''' (S-P) | ||
| + | |BBa_R0040 (we already had this plasmid at -20°C) (E-X) | ||
| + | |BBa_B0030-BBa_C0051-'''BBa_B0030'''-BBa_C0079 (4) (E-S) | ||
| + | |} | ||
| + | |||
| + | *Gel run/cut/gel extraction for all these parts. | ||
| + | |||
| + | *(We already had BBa_B1006 (E-X) and BBa_R0062 (E-X)) | ||
| + | |||
| + | *Ligation: | ||
| + | #BBa_J23100-'''BBa_B0030''' (S-P) -BBa_C0012 (X-P) (1:2 ratio instead of 1:6) | ||
| + | #'''BBa_B1006 (E-P)''' + BBa_J23100-BBa_B0030 (E-S, 1:4) -BBa_C0012 (X-P, 1:2) (we decided to try this double ligation) | ||
| + | #BBa_J23100-BBa_B0030-BBa_C0040-BBa_B1006-'''BBa_R0040''' | ||
| + | #BBa_R0051-BBa_B0030-BBa_C0062-BBa_B1006-'''BBa_R0062''' | ||
| + | #BBa_B0030-BBa_C0051-BBa_B0030-BBa_C0079-'''BBa_B1006''' | ||
| + | #BBa_B0030-BBa_I15009-'''BBa_B1006''' | ||
| + | |||
| + | *We incubated ligations at 16°C overnight. | ||
| + | |||
| + | *We infected 9 ml LB + Amp with 30 µl of BBa_R0079 glycerol stock. | ||
| + | |||
| + | <br><br> | ||
| + | '''07/12/08''' | ||
| + | <br> | ||
| + | *Glycerol stocks for: | ||
| + | {|cellpadding="20px" | ||
| + | |BBa_R0010 | ||
| + | |BBa_R0079 | ||
| + | |BBa_B0030-BBa_C0078-'''BBa_B1006''' | ||
| + | |} | ||
| + | |||
| + | *Miniprep for these 3 parts. | ||
| + | |||
| + | *We put these 3 purified plasmids at -20°C. | ||
| + | |||
| + | *We also put the 6 ovenight ligations at -20°C. Next week they will be transformed! | ||
| + | |||
<br> | <br> | ||
Latest revision as of 21:26, 26 October 2008
Notebook
| Week 1 | Week 2 | Week 3 | Week 4 | Week 5 | Week 6 | Week 7 |
|---|---|---|---|---|---|---|
| Week 8 | Week 9 | Week 10 | Week 11 | Week 12 | Week 13 | Week 14 |
| Week 15 | Week 16 | Week 17 | Week 18 | Week 19 | Week 20 | Week 21 |
| Week 22 | Week 23 | Week 24 |
Week 8: 07/7/08 - 07/12/08
07/7/08
- Colony PCR (5 colonies for each plate) for:
- BBa_J23100-BBa_B0030-BBa_C0012 (for the second time, we hoped to find true positive colonies)
- BBa_J23100-BBa_B0030-BBa_I15010 (for the second time, we hoped to find true positive colonies)
- BBa_B0030-BBa_E0040-BBa_B1006
- BBa_B0030-BBa_C0051-BBa_B0030
- BBa_B0030-BBa_E1010-BBa_B1006
- Gel results:
- No true positives for BBa_J23100-BBa_B0030-BBa_C0012
- No true positives for BBa_J23100-BBa_B0030-BBa_I15010
- Non pure true positives for BBa_B0030-BBa_E0040-BBa_B1006
- Pure true positives for BBa_B0030-BBa_C0051-BBa_B0030
- Non pure true positives for BBa_B0030-BBa_E1010-BBa_B1006
- We chose to keep the first colony for all the 3 working ligation plates.
- NOTE: BBa_B0030-BBa_E0040-BBa_B1006 and BBa_B0030-BBa_E1010-BBa_B1006 are final parts; we decided to sequence these 2 parts even if gel showed a weak contamination: sequencing results will tell us if there are false positive plasmids in those colonies or if the contamination was only a PCR contamination.
- We infected 9 ml LB + suitable antibiotic with 30 µl of these glycerol stocks:
| BBa_B0030 | BBa_I15010 | BBa_B0030-BBa_E0040-BBa_B1006 (1) | BBa_B0030-BBa_E1010-BBa_B1006 (1) |
| BBa_C0012 | BBa_R0062 | BBa_B0030-BBa_C0051-BBa_B0030 | BBa_J23100-BBa_B0030 |
- We incubated the 8 cultures at 37°C, 220 rpm overnight.
- Tomorrow we will be ready to perform NINE LIGATIONS...(@_@!)
07/8/08
- We received sequencing results for BBa_B0030-BBa_C0078: sequence showed an extra DNA fragment between BBa_C0078 and Suffix...We decided to ignore it because there was a stop codon before that fragment.
- Glycerol stocks for:
| BBa_B0030 | BBa_I15010 | BBa_B0030-BBa_E0040-BBa_B1006 (1) |
| BBa_C0012 | BBa_R0062 | BBa_B0030-BBa_C0051-BBa_B0030 |
| BBa_J23100-BBa_B0030 | BBa_B0030-BBa_E1010-BBa_B1006 (1) |
- Miniprep for these parts.
- Plasmid digestion for:
| BBa_B0030 (S-P) | BBa_B0030-BBa_E0040-BBa_B1006 (1) (E-X) | BBa_B0030-BBa_E1010-BBa_B1006 (1) (E-X) |
| BBa_C0012 (X-P) | BBa_B0030-BBa_C0051-BBa_B0030 (S-P) | BBa_J23100-BBa_B0030 (E-S) |
| BBa_I15010 (X-P) | BBa_J23100-BBa_B0030-BBa_C0040 (E-S) | BBa_R0051-BBa_B0030-BBa_C0062 (E-S) |
| BBa_R0062 (E-X) | BBa_B0030-BBa_C0061-BBa_B1006 (E-S) |
- Gel run/cut/gel extraction for:
| BBa_I15010 (X-P) | BBa_J23100-BBa_B0030 (E-S) | BBa_J23100-BBa_B0030-BBa_C0040 (E-S) |
| BBa_C0012 (X-P) | BBa_R0051-BBa_B0030-BBa_C0062 (E-S) | BBa_B0030-BBa_C0061-BBa_B1006 (E-S) |
- DNA precipitation with sodium acetate for:
| BBa_B0030 (S-P) | BBa_B0030-BBa_E0040-BBa_B1006 (1) (E-X) | BBa_B0030-BBa_E1010-BBa_B1006 (1) (E-X) |
| BBa_R0062 (E-X) | BBa_B0030-BBa_C0051-BBa_B0030 (S-P) |
- (We already had BBa_I15009(X-P) and BBa_B1006(E-X))
- Ligations:
- BBa_B0030-BBa_I15009
- BBa_J23100-BBa_B0030-BBa_C0040-BBa_B1006
- BBa_R0051-BBa_B0030-BBa_C0062-BBa_B1006
- BBa_B0030-BBa_C0051-BBa_B0030-BBa_C0079
- BBa_B0030-BBa_C0061-BBa_B1006-BBa_R0062
- BBa_J23100-BBa_B0030-BBa_C0012 (again)
- BBa_J23100-BBa_B0030-BBa_I15010 (again)
- BBa_J23100-BBa_B0030-BBa_E0040-BBa_B1006 (to test the part)
- BBa_J23100-BBa_B0030-BBa_E1010-BBa_B1006 (to test the part)
- We incubated ligations at 16°C overnight.
- We infected 9 ml LB + Amp with 30 µl of BBa_B1006 and BBa_B0030-BBa_C0078 glycerol stocks. We incubated the 2 cultures at 37°C, 220 rpm ovenight.
07/9/08
- We transformed the 9 ligations (2 µl) and plated transformed bacteria. We incubated plates at 37°C ovenight.
- Glycerol stocks for BBa_B1006 and BBa_B0030-BBa_C0078.
- Miniprep for BBa_B1006 and BBa_B0030-BBa_C0078.
- Plasmid digestion:
| BBa_B1006 (E-X) | BBa_B0030-BBa_C0078 (E-S) |
- Gel run/cut/gel extraction for BBa_B0030-BBa_C0078 (E-S).
- DNA precipitation with sodium acetate for BBa_B1006 (E-X).
- Ligation: BBa_B0030-BBa_C0078-BBa_B1006
- We incubated ligation at 16°C overnight.
07/10/08
- We transformed BBa_B0030-BBa_C0078-BBa_B1006 ligation (2 µl) and plated transformed bacteria. We incubated the plate at 37°C ovenight.
- All the ligation plates showed colonies! There were carpets, but we could pick up some single colonies for colony PCR.
- Colony PCR for:
- BBa_B0030-BBa_I15009
- BBa_J23100-BBa_B0030-BBa_C0040-BBa_B1006
- BBa_R0051-BBa_B0030-BBa_C0062-BBa_B1006
- BBa_B0030-BBa_C0051-BBa_B0030-BBa_C0079
- BBa_B0030-BBa_C0061-BBa_B1006-BBa_R0062
- BBa_J23100-BBa_B0030-BBa_C0012
- BBa_J23100-BBa_B0030-BBa_I15010
- Gel results:
- BBa_B0030-BBa_I15009 1st colony was chosen.
- BBa_J23100-BBa_B0030-BBa_C0040-BBa_B1006 5th colony was chosen.
- BBa_R0051-BBa_B0030-BBa_C0062-BBa_B1006 2nd colony was chosen.
- BBa_B0030-BBa_C0051-BBa_B0030-BBa_C0079 4th colony was chosen.
- BBa_B0030-BBa_C0061-BBa_B1006-BBa_R0062 3rd colont was chosen.
- BBa_J23100-BBa_B0030-BBa_C0012 did not show true positive colonies.
- BBa_J23100-BBa_B0030-BBa_I15010 did not show true positive colonies.
- We streaked BBa_J23100-BBa_B0030-BBa_E0040-BBa_B1006 and BBa_J23100-BBa_B0030-BBa_E1010-BBa_B1006 with a top to test the parts (NOTE: we could see some red colonies on BBa_J23100-BBa_B0030-BBa_E1010-BBa_B1006 plate!these colonies correspond to bacteria with correctly ligated plasmid, while normal color colonies correspond to bacteria with BBa_B0030-BBa_E1010-BBa_B1006 plasmid, without constitutive promoter).
- We infected 100 µl LB + Amp with the tips and incubated the culture at 37°C, 220 rpm for 2 hours.
- Then we watched green, blue and red fluorescence channels at microscope (50 µl of each culture):
- LB preparation: 0.5 l LB + Amp for plates.
- BBa_R0010 resuspension from Registry of Standard Parts.
- We transformed (4 µl) BBa_R0010 and plated transformed bacteria. We incubated BBa_R0010 plate at 37°C overnight.
- We infected 9 ml LB + suitable antibiotic with 30 µl of these glycerol stocks:
| BBa_C0012 | BBa_B0030-BBa_I15009 (1) |
| BBa_J23100-BBa_B0030-BBa_C0040-BBa_B1006 (5) | BBa_R0051-BBa_B0030-BBa_C0062-BBa_B1006 (2) |
| BBa_J23100-BBa_B0030 | BBa_B0030-BBa_C0051-BBa_B0030-BBa_C0079 (4) |
| BBa_B0030-BBa_C0061-BBa_B1006-BBa_R0062 (3) |
- We also picked up one colony from BBa_I15010 plate with a tip and infected 9 ml LB + Kan. We incubated the 8 cultures at 37°C, 220 rpm overnight.
07/11/08
- BBa_R0010 plate showed 2 colonies. We picked up one colony and infected 9 ml LB + Amp to grow an overnight culture.
- BBa_B0030-BBa_C0078-BBa_B1006 plate showed many colonies. We used 7 colonies to perform colony PCR.
- Gel results: all the 7 colonies contained ligated plasmid; we chose the 5th colony to grow a 9 ml overnigth culture because 5th it showed no impurities.
- Glycerol stocks for:
| BBa_C0012 | BBa_B0030-BBa_I15009 (1) |
| BBa_I15010 | BBa_J23100-BBa_B0030-BBa_C0040-BBa_B1006 (5) |
| BBa_J23100-BBa_B0030 | BBa_R0051-BBa_B0030-BBa_C0062-BBa_B1006 (2) |
| BBa_B0030-BBa_C0051-BBa_B0030-BBa_C0079 (4) | BBa_B0030-BBa_C0061-BBa_B1006-BBa_R0062 (3) |
- Miniprep for these 8 parts.
- We sent BBa_I15010 and BBa_B0030-BBa_C0061-BBa_B1006-BBa_R0062 (3) to Primm for sequencing.
- Plasmid digestion for:
| BBa_C0012 (X-P) | BBa_B0030-BBa_I15009 (1) (E-S) | BBa_B1006 (we already had this plasmid at -20°C) (E-P) |
| BBa_J23100-BBa_B0030 (E-S) | BBa_J23100-BBa_B0030-BBa_C0040-BBa_B1006 (5) (E-S) | BBa_R0051-BBa_B0030-BBa_C0062-BBa_B1006 (2) (E-S) |
| BBa_J23100-BBa_B0030 (S-P) | BBa_R0040 (we already had this plasmid at -20°C) (E-X) | BBa_B0030-BBa_C0051-BBa_B0030-BBa_C0079 (4) (E-S) |
- Gel run/cut/gel extraction for all these parts.
- (We already had BBa_B1006 (E-X) and BBa_R0062 (E-X))
- Ligation:
- BBa_J23100-BBa_B0030 (S-P) -BBa_C0012 (X-P) (1:2 ratio instead of 1:6)
- BBa_B1006 (E-P) + BBa_J23100-BBa_B0030 (E-S, 1:4) -BBa_C0012 (X-P, 1:2) (we decided to try this double ligation)
- BBa_J23100-BBa_B0030-BBa_C0040-BBa_B1006-BBa_R0040
- BBa_R0051-BBa_B0030-BBa_C0062-BBa_B1006-BBa_R0062
- BBa_B0030-BBa_C0051-BBa_B0030-BBa_C0079-BBa_B1006
- BBa_B0030-BBa_I15009-BBa_B1006
- We incubated ligations at 16°C overnight.
- We infected 9 ml LB + Amp with 30 µl of BBa_R0079 glycerol stock.
07/12/08
- Glycerol stocks for:
| BBa_R0010 | BBa_R0079 | BBa_B0030-BBa_C0078-BBa_B1006 |
- Miniprep for these 3 parts.
- We put these 3 purified plasmids at -20°C.
- We also put the 6 ovenight ligations at -20°C. Next week they will be transformed!
 "
"