|
|
| (10 intermediate revisions not shown) |
| Line 1: |
Line 1: |
| - | '''Ucp1'''
| + | <html><link rel="stylesheet" href="https://2008.igem.org/wiki/index.php?title=User:Joadelas/valencia.css&action=raw&ctype=text/css" type="text/css"></html> |
| | + | __NOTOC__ |
| | + | {{Valencia/Menu}} |
| | + | {{Valencia/Menu_Parts}} |
| | | | |
| | + | <div class="valenciaMain"> |
| | | | |
| - | '''General information'''
| + | <div style=" width:94%; margin: 0 auto;"> |
| | + | ==Construction of Valencia Team Biobricks== |
| | | | |
| - | Ucp1 is a gene which encodes for UCP1 protein.
| + | ===Preparing inserts=== |
| | | | |
| - | The uncoupling protein UCP1 is a proton carrier characteristic of brown adipose tissue.
| + | Total DNA was extracted from our yeast strains.<br> |
| - | UCP1 uncouples the respiratory chain of ATP production, converting the metabolic energy in heat.
| + | UCP-1 was amplified by PCR using oligonucleotides matching the sequence and bearing the appropriate Biobrick prefix and suffix.<br> |
| | | | |
| - | <center>
| |
| - | {|
| |
| - | |[[Image:function.jpg|400px]]
| |
| - | |}
| |
| - | </center>
| |
| | | | |
| | + | Primers' sequences (EcoRI and PstI sites in bold):<br> |
| | + | Forward: 5' '''gAATTC'''gCggCCgCTTCTAgATggTgAgTTCgACAACTTC 3';<br> |
| | + | Reverse: 5' TACTAgTAgCggCCg'''CTgCAg'''CTATgTggTgCAgTCCACTg 3' <br> |
| | + | |
| | | | |
| - | UCP1 is a 33kd protein which is exclusively located in the brown adipocites.
| + | PCR was conducted as follows:<br> |
| - | It is an integral protein present in inner mitocondrial membrane.
| + | |
| | | | |
| - | The protein has a tripartite structure. The structure displays an around 100 residues region which is three times repeated. Each part encodes for two transmembrane segments and one long hydrophilic loop.The functional carrier unit is an homodimer.
| + | <ol>A first denaturation cycle |
| | + | <ol>94º 3'</ol> |
| | + | Followed by 30 amplification cycles: |
| | + | <ol>94º 30'' |
| | + | 60º 1' 30'' |
| | + | 72º 1'</ol> |
| | + | And a final extension step: |
| | + | <ol>72º 10'</ol> |
| | + | </ol> |
| | | | |
| - | <center>
| |
| - | {|
| |
| - | |[[Image:Structure.jpg|400px]]
| |
| - | |}
| |
| - | </center>
| |
| | | | |
| | + | '''Results:'''<br> |
| | | | |
| - | The main difference between UCP1 and most of the proteins with a nuclear codification is the lack of the importation targeting to the mitochondria in UCP+ proteins.
| + | [[Image:Valencia_PCRresults.jpg]]<br> |
| - | The condition that determines the mitochondria as the protein target lays in the first loop which protudes in the mitochondrial matrix.
| + | Intense amplicons, of the expected size, (about 1 kb) were produced for UCP-1, 175-deleted and 76 deleted. <br> |
| | + | Amplicons were digested (H buffer) with EcoRI y PstI.<br> |
| | | | |
| - | The second loop of the matrix is essential for the insertion of the protein in the inner mitochondrial matrix.
| + | ===Preparing vectors=== |
| - | Purine nucleotides act as inhibitors of protein activity and esterificated fatty acids act as inductors.
| + | |
| | + | Competent cells were transformed with: pSB1AK3 and pUC18 plasmids <br> |
| | | | |
| | | | |
| | + | Plasmids were extracted (High pure miniprep plasmid isolation kit ROCHE) <br> |
| | + | Plasmid were digested with EcoRI and PstI<br> |
| | | | |
| - | '''Sequence'''
| |
| | | | |
| | + | ===Ligating Biobricks into plasmids=== |
| | | | |
| - | [[Image:secuencia UCP1.jpg]]
| + | Both plasmids and inserts were run into 0.8% 0.5X TBE agarose gels and DNA bands excised with a clean scalpel. DNA was extracted from agarose blocks (ultra clean gel spin, DNA purification Kit, MO BIO laboratories).<br> |
| | + | T4 Ligase was used to ligate inserts and vectors for 1 h at room temperature (2X quick buffer was used).<br> |
| | + | Competent cells were transformed and resulting colonies (Amp LB) screened with Fw and Rv primers to confirm the presence of inserts. <br> |
| | + | pSB1AK3 containing UCP-1, 175-deleted and 76-deleted were sent to the Registry <br> |
| | | | |
| | | | |
| | | | |
| - | '''UCP 175 deleted'''
| |
| | | | |
| | | | |
| - | | + | </div> |
| - | The uncoupling protein UCP 175 deleted is a proton carrier which is obtaines from the direct mugenesis of the Ucp1 gene.
| + | </div> |
| - | Ucp 175 has a deletion in the triplet the encodes for the Glycine 175.
| + | |
| - | The mutant shows a generation time and an heat up capacity higher than UCP1.
| + | |
| - | UCP 175 deleted uncouples the respiratory chain of ATP production, converting the metabolic energy in heat.
| + | |
| - | The mutant shows a generation time and an heat up capacity higher than UCP1.
| + | |
| - | | + | |
| - | <center>
| + | |
| - | {|
| + | |
| - | |[[Image:function.jpg|400px]]
| + | |
| - | |}
| + | |
| - | </center> | + | |
| - | | + | |
| - | | + | |
| - | UCP 175 deleted is a 33kd protein.
| + | |
| - | The protein has a tripartite structure. The structure displays an around 100 residues region which is three times repeated. Each part encodes for two transmembrane segments and one long hydrophilic loop.The functional carrier unit is an homodimer.
| + | |
| - | | + | |
| - | <center>
| + | |
| - | {|
| + | |
| - | |[[Image:Structure.jpg|400px]]
| + | |
| - | |}
| + | |
| - | </center> | + | |
| - | | + | |
| - | | + | |
| - | The main difference between UCP 175 deleted and most of the proteins with a nuclear codification is the lack of the importation targeting to the mitochondria in UCP 175 deleted proteins.
| + | |
| - | | + | |
| - | The condition that determines the mitochondria as the protein target lays in the first loop which protudes in the mitochondrial matrix.
| + | |
| - | | + | |
| - | The second loop of the matrix is essential for the insertion of the protein in the inner mitochondrial matrix.
| + | |
| - | | + | |
| - | That protein is without direct regulation and its generation time depends on the inner activity the protein.
| + | |
| - | | + | |
| - | | + | |
| - | | + | |
| - | '''Sequence:'''
| + | |
| - | | + | |
| - | [[Image:Sequence UCP 175 deleted.jpg]]
| + | |
| - | | + | |
| - | ''' Graphic about the growth of four different strains of Saccharomyces cerevisiae transformed with four kinds of UCP genes:'''
| + | |
| - | | + | |
| - | | + | |
| - | [[Image:Creixement.jpg]]
| + | |
| - | | + | |
| - | Y axis = Population in OD (optic density) units.
| + | |
| - | X axis = Time in hours.
| + | |
| - | | + | |
| - | | + | |
| - | | + | |
| - | '''UCP 76 deleted'''
| + | |
| - | | + | |
| - | | + | |
| - | | + | |
| - | The uncoupling protein UCP 76 deleted is a proton carrier which is obtaines from the direct mugenesis of the Ucp1 gene.
| + | |
| - | Ucp 76 has a deletion in the triplet the encodes for the Glycine 76.
| + | |
| - | The mutant shows a generation time and an heat up capacity higher than UCP1.
| + | |
| - | UCP 76 deleted uncouples the respiratory chain of ATP production, converting the metabolic energy in heat.
| + | |
| - | The mutant shows a generation time and an heat up capacity higher than UCP1.
| + | |
| - | | + | |
| - | | + | |
| - | <center>
| + | |
| - | {|
| + | |
| - | |[[Image:function.jpg|400px]]
| + | |
| - | |}
| + | |
| - | </center>
| + | |
| - | | + | |
| - | | + | |
| - | UCP 76 deleted is a 33kd protein.
| + | |
| - | The protein has a tripartite structure. The structure displays an around 100 residues region which is three times repeated. Each part encodes for two transmembrane segments and one long hydrophilic loop.The functional carrier unit is an homodimer.
| + | |
| - | | + | |
| - | <center>
| + | |
| - | {|
| + | |
| - | |[[Image:Structure.jpg|400px]]
| + | |
| - | |}
| + | |
| - | </center>
| + | |
| - | | + | |
| - | | + | |
| - | The main difference between UCP 76 deleted and most of the proteins with a nuclear codification is the lack of the importation targeting to the mitochondria in UCP 76 deleted proteins.
| + | |
| - | | + | |
| - | The condition that determines the mitochondria as the protein target lays in the first loop which protudes in the mitochondrial matrix.
| + | |
| - | | + | |
| - | The second loop of the matrix is essential for the insertion of the protein in the inner mitochondrial matrix.
| + | |
| - | | + | |
| - | That protein is without direct regulation and its generation time depends on the inner activity the protein.
| + | |
| - | | + | |
| - | | + | |
| - | | + | |
| - | '''Sequence:'''
| + | |
| - | | + | |
| - | [[Image:Sequence UCP 76 deleted.jpg]]
| + | |
| - | | + | |
| - | ''' Graphic about the growth of four different strains of Saccharomyces cerevisiae transformed with four kinds of UCP genes:'''
| + | |
| - | | + | |
| - | | + | |
| - | [[Image:Creixement.jpg]]
| + | |
| - | | + | |
| - | Y axis = Population in OD (optic density) units.
| + | |
| - | X axis = Time in hours.
| + | |
Construction of Valencia Team Biobricks
Preparing inserts
Total DNA was extracted from our yeast strains.
UCP-1 was amplified by PCR using oligonucleotides matching the sequence and bearing the appropriate Biobrick prefix and suffix.
Primers' sequences (EcoRI and PstI sites in bold):
Forward: 5' gAATTCgCggCCgCTTCTAgATggTgAgTTCgACAACTTC 3';
Reverse: 5' TACTAgTAgCggCCgCTgCAgCTATgTggTgCAgTCCACTg 3'
PCR was conducted as follows:
A first denaturation cycle
94º 3'
Followed by 30 amplification cycles:
94º 30
60º 1' 30
72º 1'
And a final extension step:
72º 10'
Results:
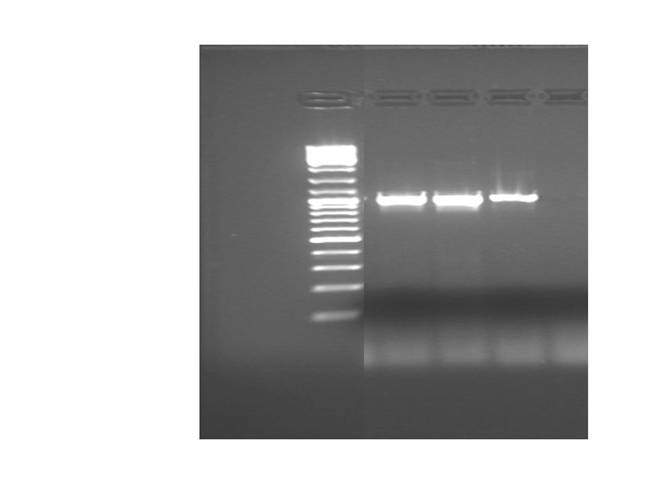
Intense amplicons, of the expected size, (about 1 kb) were produced for UCP-1, 175-deleted and 76 deleted.
Amplicons were digested (H buffer) with EcoRI y PstI.
Preparing vectors
Competent cells were transformed with: pSB1AK3 and pUC18 plasmids
Plasmids were extracted (High pure miniprep plasmid isolation kit ROCHE)
Plasmid were digested with EcoRI and PstI
Ligating Biobricks into plasmids
Both plasmids and inserts were run into 0.8% 0.5X TBE agarose gels and DNA bands excised with a clean scalpel. DNA was extracted from agarose blocks (ultra clean gel spin, DNA purification Kit, MO BIO laboratories).
T4 Ligase was used to ligate inserts and vectors for 1 h at room temperature (2X quick buffer was used).
Competent cells were transformed and resulting colonies (Amp LB) screened with Fw and Rv primers to confirm the presence of inserts.
pSB1AK3 containing UCP-1, 175-deleted and 76-deleted were sent to the Registry

 "
"