Team:Freiburg/Project
From 2008.igem.org
m |
|||
| Line 39: | Line 39: | ||
<tr> | <tr> | ||
<td> | <td> | ||
| - | [[Team:Freiburg/ | + | [[Team:Freiburg/Modeling|Modeling]]<br> |
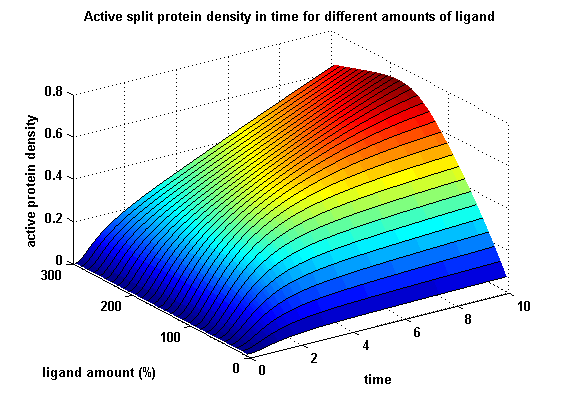
| - | + | The dimerization of the extracellular receptor domains is a important necessity for the functionality of our Modular Synthetic Receptor System. Presenting the system a stimulus in the form of spatial arranged ligands (nitro-iodo-phenol or fluorescein molecules), the extracellular domains dimerize, thus the corresponding intracellular parts such as the split lactamase halves or split fluorescent proteins complement to measureable output. To analyse the theoretical functionality due to dimerization, two receptor dimerization models (one T cell receptor model and one general receptor model) are introduced and discussed and a proper model for the Modular Synthetic Receptor System is constructed. It consists of 10 reaction kinetic equations, 9 ordinary differential equations including 25 variable parameters. Matlab m-files are embedded for further inspection. | |
</td> | </td> | ||
| + | <td> | ||
| + | [[image:Freiburg2008_MSRSdNN.png|thumb|right|200px|Split protein activity in time dependent on ligand amount]]<br> | ||
| + | </td> | ||
| + | </tr> | ||
| + | </table> | ||
| + | ---- | ||
| + | <table> | ||
| + | <tr> | ||
<td> | <td> | ||
[[Image:Team-Freiburg2008_Lipo_alpha_nCFP.png|thumb|right|200 px|structural model of expression part Bba_K157037]] | [[Image:Team-Freiburg2008_Lipo_alpha_nCFP.png|thumb|right|200 px|structural model of expression part Bba_K157037]] | ||
| + | </td> | ||
| + | <td> | ||
| + | [[Team:Freiburg/3D-Modeling|3D-Modeling]]<br> | ||
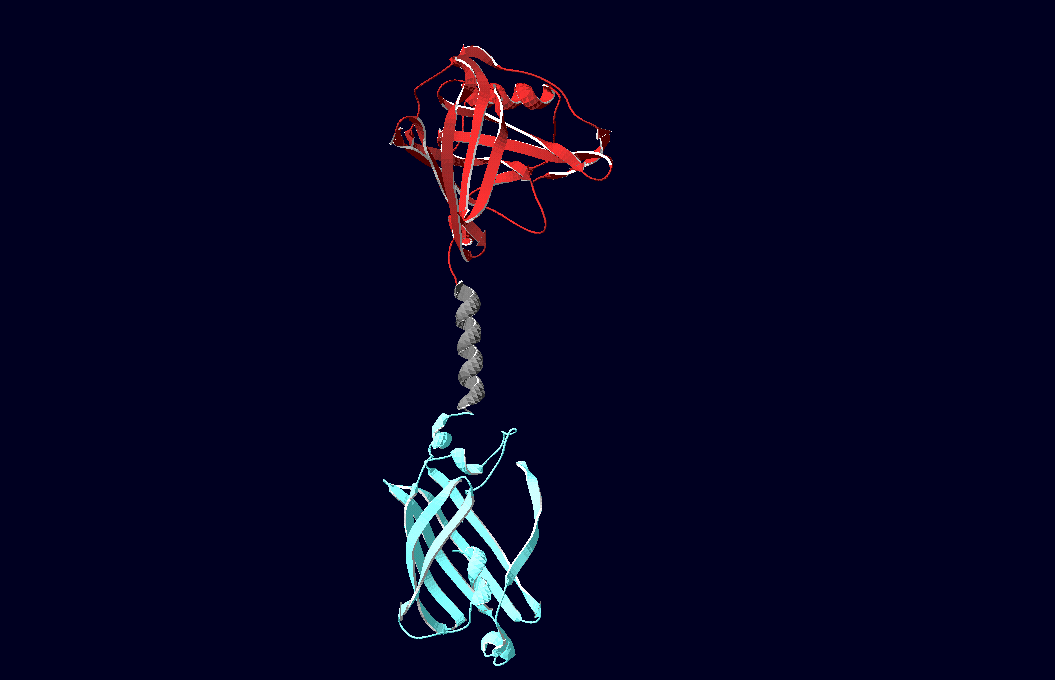
| + | We have created various three-dimensional models of our constructs using Pymol and SwissPdbViewer. Pdb-files of the huge DNA-Origami molecule were created as adequate as possible using Nano-Engineer version 1.0 and then edited in Pymol. These Models were used to plan the spatial arrangement and, thus, input-pattern of antigens at a nanometer scale. | ||
</td> | </td> | ||
</tr> | </tr> | ||
| Line 51: | Line 63: | ||
<tr> | <tr> | ||
<td> | <td> | ||
| - | [[ | + | [[DNA-Origami|DNA-Origami]]<br> |
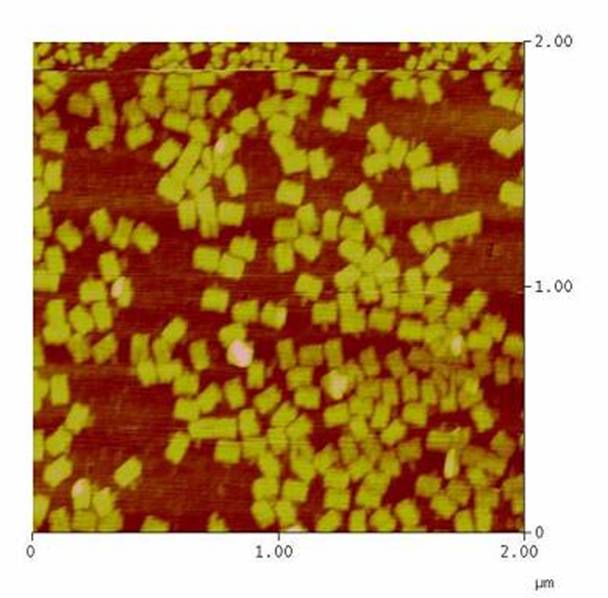
| + | We have created various three-dimensional models of our constructs using Pymol and SwissPdbViewer. Pdb-files of the huge DNA-Origami molecule were created as adequate as possible using Nano-Engineer version 1.0 and then edited in Pymol. These Models were used to plan the spatial arrangement and, thus, input-pattern of antigens at a nanometer scale. | ||
</td> | </td> | ||
<td> | <td> | ||
| - | [[Team | + | [[Image:Team Freiburg2008-Origami 1zu5.jpg|thumb|right|200 px|AFM-measurement of DNA-Origami]] |
| - | + | ||
</td> | </td> | ||
</tr> | </tr> | ||
</table> | </table> | ||
| - | |||
---- | ---- | ||
<table> | <table> | ||
<tr> | <tr> | ||
<td> | <td> | ||
| - | [[ | + | [[Image:Freiburg2008 Konstrukte.jpg|thumb|right|200px|cloning scheme]] |
| - | + | ||
| - | + | ||
</td> | </td> | ||
<td> | <td> | ||
| - | [[ | + | [[Team:Freiburg_Cloning Strategy|Cloning Strategy]]<br> |
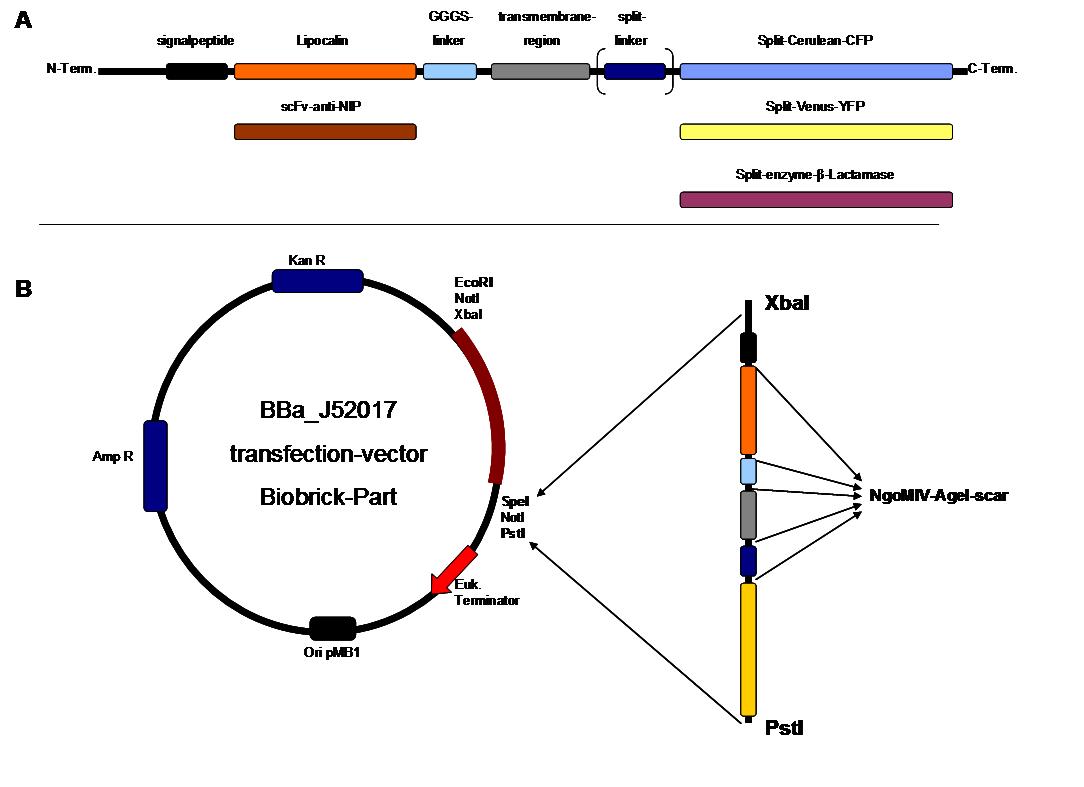
| + | All of our constructs were cloned successfully using our extended pre- and suffix and plasmid "pMA" (part Bba_K157000). The broad combinatorial range given by the modular concept was almost fully exploited, so that we ended up with the submission of 13 basic and 28 composite parts. Once a construct was completed, it was cloned into the CMV-promoted transfection vector (part Bba_K157040) and used for transfection of 293T-cells. | ||
</td> | </td> | ||
</tr> | </tr> | ||
</table> | </table> | ||
| - | |||
| - | |||
---- | ---- | ||
<table> | <table> | ||
<tr> | <tr> | ||
<td> | <td> | ||
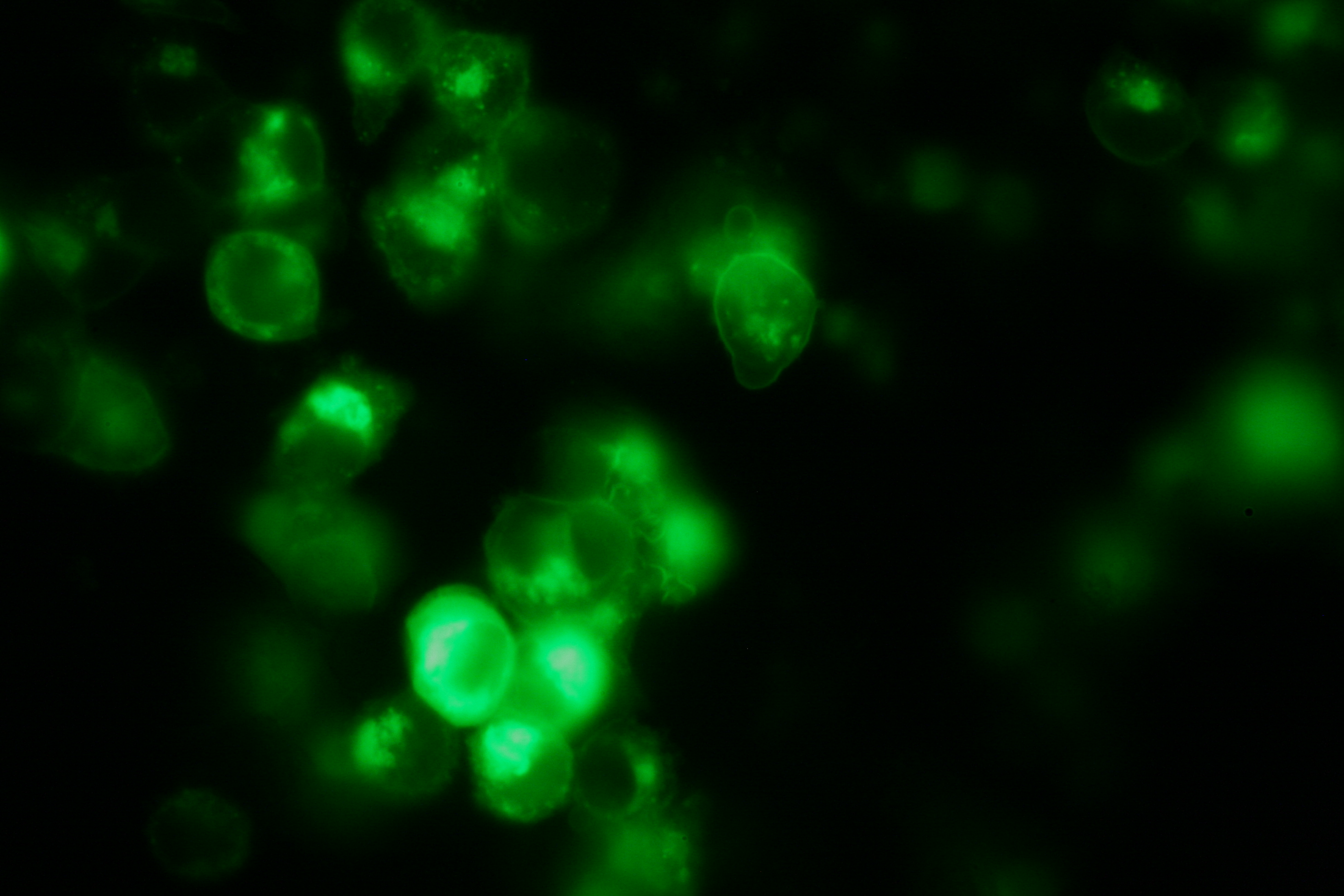
| - | [[ | + | [[Image:Freiburg2008_Fluomembrane.jpg|thumb|right|200px|membrane-localization of Part Bba_K157032-YFP]] |
</td> | </td> | ||
<td> | <td> | ||
| - | [[ | + | [[Team:Freiburg_Transfection and Synthetic Receptor|Transfection and Synthetic Receptor Activation]]<br> |
| + | Transfection of 293T-cells has also been shown to work for most of our fusion proteins; so far, we could even show that at least our constructs with lipocalin FluA as extracellular domaine are integrated into the membrane.<br> | ||
| + | All transfections were carried out with part Bba_K157040, one of our composite parts consisting of the transfection vector Bba_J52017 and a CMV-promotor. Due to the limited time range we were not able to activate/dimerize any of the various possible "receptor"-pairs yet, but we are hoping to receive a positive result in that concern until the jamboree. | ||
</td> | </td> | ||
</tr> | </tr> | ||
</table> | </table> | ||
| - | |||
---- | ---- | ||
<table> | <table> | ||
<tr> | <tr> | ||
<td> | <td> | ||
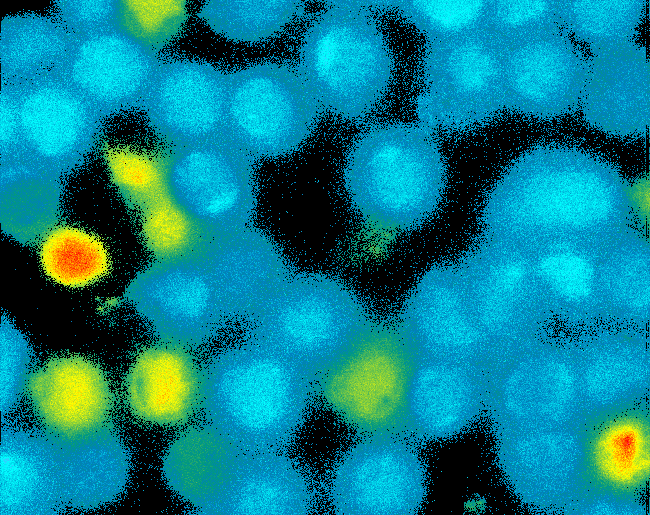
| - | [[Team: | + | [[Team:Freiburg_Calcium Imaging|Cell Stability, Ca2+ Signaling and DNA-Origami-Binding]] |
| - | + | ||
</td> | </td> | ||
<td> | <td> | ||
| - | [[Image: | + | [[Image:TeamFreiburg2008_Pervanadate1.png|thumb|right|200px|Ca-influx measurement]] |
</td> | </td> | ||
</tr> | </tr> | ||
</table> | </table> | ||
| + | |||
| + | |||
Revision as of 19:38, 29 October 2008
|
Project Report |
_project report
IntroductionThis year´s main project is the attempt to create an "artificial receptor-system", featuring extra- and intracellular modules as well as suitable transmembrane regions.
The intracellular domaine of our receptor-device is build by halves of split reporter-proteins that can reassemble and will then produce readable output, e. g. fluorescence.
Each one of these protein-halves is connected to its extracellular domaine by a single-span transmembrane-helix.
The extracellular or detecting domaine consists of a protein or peptide with the ability to bind a certain molecule. SubprojectsModeling Highlights
LiteratureSplit-fluorophores: |
 "
"