Team:Bologna
From 2008.igem.org
Rita.morini (Talk | contribs) |
|||
| Line 42: | Line 42: | ||
|titolo=[[Team:Bologna/Team|TEAM]] | |titolo=[[Team:Bologna/Team|TEAM]] | ||
|contenuto=[[Image:TeamBo2.JPG|250px|right]] | |contenuto=[[Image:TeamBo2.JPG|250px|right]] | ||
| - | The leading idea behind | + | The leading idea behind the UniBO Team was to put in touch students of the same University with different backgrounds to favor competence and skill sharing. Students from different Faculties (Biotechnology, Pharmacy and Engineering) answered to the iGEM call and formed the TEAM. They were moved by a common desire for new experiences and they decided to participate in this exciting competition to go deep in Synthetic Biology. The Team worked hard during the summer to become fully operative, overcoming differences both in training and personality. |
<br><br> | <br><br> | ||
[[Team:Bologna/Team|Go to the page!]] | [[Team:Bologna/Team|Go to the page!]] | ||
Revision as of 00:26, 30 October 2008
| HOME | PROJECT | TEAM | SOFTWARE | MODELING | WET LAB | LAB-BOOK | SUBMITTED PARTS | BIOSAFETY AND PROTOCOLS |
|---|
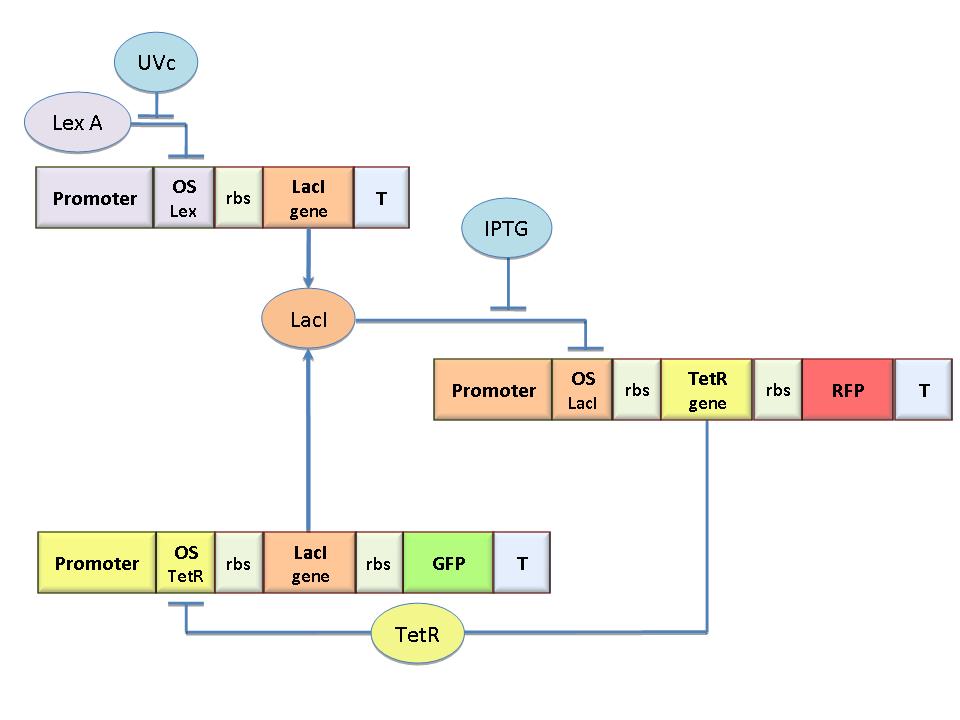
Our project aims to design a bacterial reprogrammable binary memory (EPROM) with genetically engineered E.coli. To engineer bacteria we designed a genetic Flip-Flop formed by an epigenetic memory sensitive to IPTG - for memory reset- and an UV-sensitive trigger to set the memory ON. We designed the Flip-Flop by model-based analysis. The core elements are two mutually regulated promoters. Each of them is composed of a constitutive promoter and an independent operator sequence. Thus, transcriptional strength and repressor binding affinity can be independently fixed. Since operator sites are still lacking in the Registry as standard parts, we cloned operator sequences for LacI, TetR, Lambda and LexA repressors and established an experimental procedure to characterize them. These parts allow the rational design of regulated promoters and we expect them to be a benefit in many Synthetic Biology applications.
Click here for more information...
The leading idea behind the UniBO Team was to put in touch students of the same University with different backgrounds to favor competence and skill sharing. Students from different Faculties (Biotechnology, Pharmacy and Engineering) answered to the iGEM call and formed the TEAM. They were moved by a common desire for new experiences and they decided to participate in this exciting competition to go deep in Synthetic Biology. The Team worked hard during the summer to become fully operative, overcoming differences both in training and personality.
Go to the page!
The aim of this work area is to determine the characteristics of the promoters that form the circuit. In this section are also collected a brief description of our model, all the equations that describe analytically our circuit, the most meaningful graphics and the results of simulations. Enjoy yourself!
Into this section you can find two software tools developed in our lab for this competition and that we would like to share with other Teams. One tool is a bacteria fluorescence image analyzer that segments the bacteria, counts their number, computes the size and the fluorescence intensity for each segmented bacterium. The second tool searches data in the Registry automatically to find parts by entry a part name or a string with short part description or a nucleotide sequence. You can download them from this page.
To obtain regulated promoters we synthesized four libraries of operator sequences, respectively for LacI, TetR, cI and LexA repressor proteins. Here is how we designed the operator libraries, how we isolated single operators and try to clone them in the standard format and how we used operators to tune promoter activation in response to UV induction.So wet!
Aknowledgements
Our Team is funded by:
- [http://www.unibo.it/Portale/default.htm University of Bologna]
- [http://serinar.criad.unibo.it Ser.In.Ar. Cesena]
 "
"