Team:ESBS-Strasbourg/System construction
From 2008.igem.org
(→Balance of R and cA) |
|||
| (9 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
<div id="header">{{Template:Team:ESBS-Strasbourg/Templates/Header}}</div> | <div id="header">{{Template:Team:ESBS-Strasbourg/Templates/Header}}</div> | ||
| - | <div id="maincontent" style="margin-top: | + | <div id="maincontent" style="margin-top:150px;"> |
| + | __notoc__ | ||
| + | =System construction= | ||
| + | ==Goal== | ||
| + | We intend to construct a cell-cycle dependent toggle switch in eucaryotic cells. This system can then be used as the basis for a binary cell division counter. | ||
| - | + | ==Organism== | |
| - | + | ''Saccharomyctes cerevisiae'' is in respect to the cell cycle regulation the best understood model organism. This will give us the possibility to develop our project with high accuracy on a theoretical level. | |
| + | Furthermore, generation times are in the right order to be able to express and also to degrade proteins within one generation time in a predictable manner. There is also the advantage of features like easy transformation and high rates of homologous recombination. In turn, the eucaryotic nature of yeast will allow an easy transfer of the system to more complex eucaryots as for example HeLa cells. | ||
| - | = | + | ==Counting== |
| - | <br> | + | The mode of binary counting appeared more elegant to us than the continuous way of counting, because the former would need less reporter compounds. So we decided to implement a binary counting system. Thus, this system would allow us to count to a higher number because of the current limitations of reporter constructs. Secondly, the binary counting mechanism would allow us to construct each bit in a modular manner, hence all further bits could be easily attached through minor changes in this standard bit module, the cell cycle dependent toggle switch. |
| + | |||
| + | ==BioBrick Standard== | ||
| + | In order to keep the interference between our system and the host metabolism to a minimum we are envisaging the use of yeast parts only where an interaction with the host is unavoidable. There are two unavoidable interfaces which are the dependence on a cell cycle specific signal as input and the use of the host transcriptional and translational machinery to ensure the expression of the system. For all other components we are planning to revert to the vast quantity of procaryotic parts from the Registry. Thus, our project will include various fusion proteins of mixed (eukaryotic, prokaryotic) nature.<br> | ||
| + | By regarding each domain as a part on its own right a rapid construction of components (for example: TFs and Repressors) with different properties would be possible (see also [https://2008.igem.org/Team:ESBS-Strasbourg/Essential_publications Dhar, PK; 2007]). However, it is not be possible to construct fusion proteins by using the BioBrick Standard developed by Knight ([https://2008.igem.org/Team:ESBS-Strasbourg/Essential_publications Knight, T; 2003]) since the 8 base between two parts would lead to a frame shift. Therefore, we envisage the use of the standard developed by Silver ([https://2008.igem.org/Team:ESBS-Strasbourg/Essential_publications Philips, IE; Silver, PA; 2007]) having only six base pairs between two parts. The transformation of existing parts into the Silver standard will therefore be an important part of our project.<br> | ||
| + | In respect to the minimal interference between the system and the host one could even go further by introducing also the whole protein expression machinery from prokaryots. We termed such an approach the [https://2008.igem.org/Team:ESBS-Strasbourg/Project_idea_history e-language]. | ||
| + | |||
| + | ==Architecture== | ||
| + | ===System level=== | ||
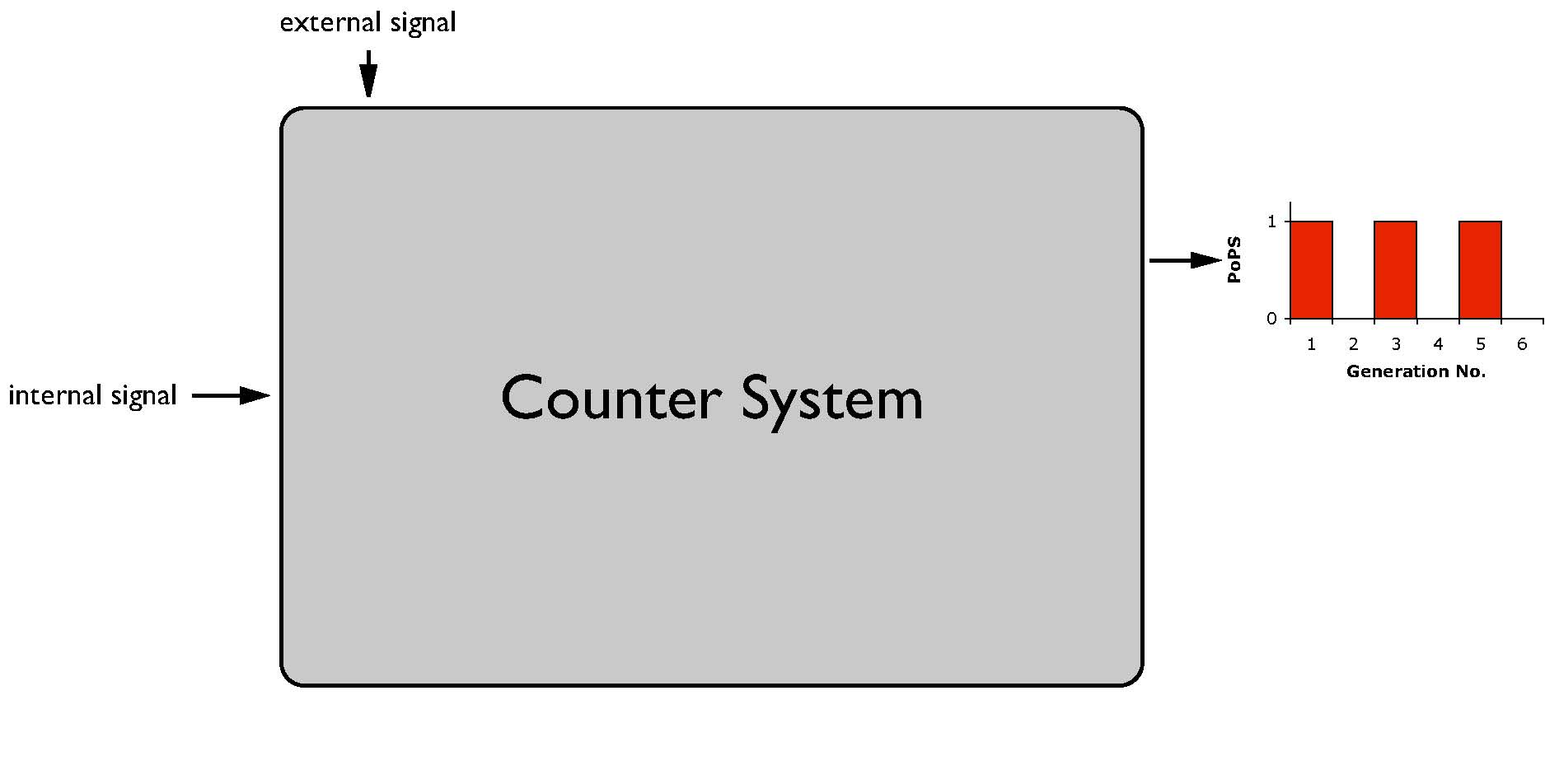
| + | [[Image:ESBS_Device1.jpg|thumb|right|255px|System level]] | ||
| + | Technical spoken, our goal is to develop a system which, once activated through an external signal, gives us alternately a PoPS output, no output respectively (see figure System level). This system is equal to the first bit of the possible cell division counter, thus would allow us to count to one. According to the idea of synthetic biology, one can use this system by using the adequate PoPS signal without a further understanding of the system's functioning. Thus, the system is available whenever a cell cycle dependent switch is needed. | ||
| + | |||
| + | ===Device level=== | ||
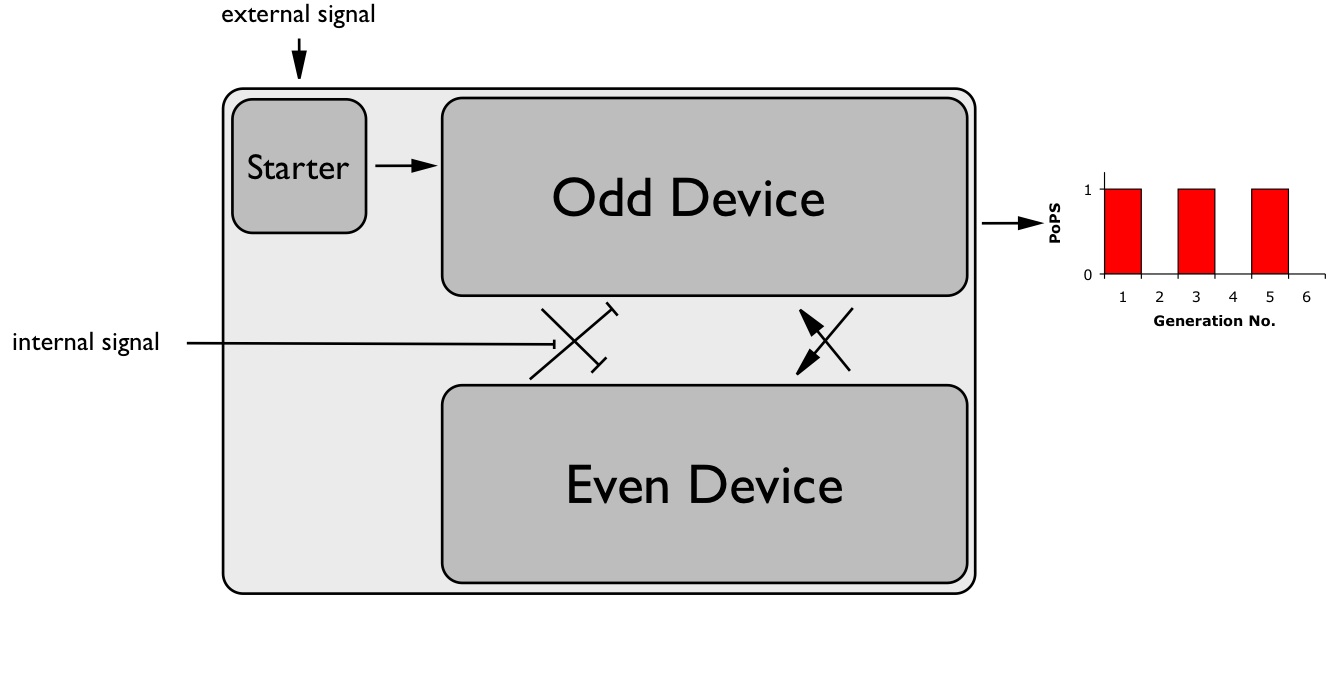
| + | [[Image:ESBS_Device2.jpg|thumb|right|255px|Device level]] | ||
| + | The figure on the right is showing our construct architecture on a device level. One can appreciate the functional units of our system. The two devices, odd device and even device, will be alternatively activated in successive generations. By coupling the PoPS outcome of the even device we will obtain the wished expression pattern. Additionally to these two devices, a starter device for the integration of the external start signal and a reporter device (not shown) will be necessary to get constructed. | ||
| + | |||
| + | ===Parts level=== | ||
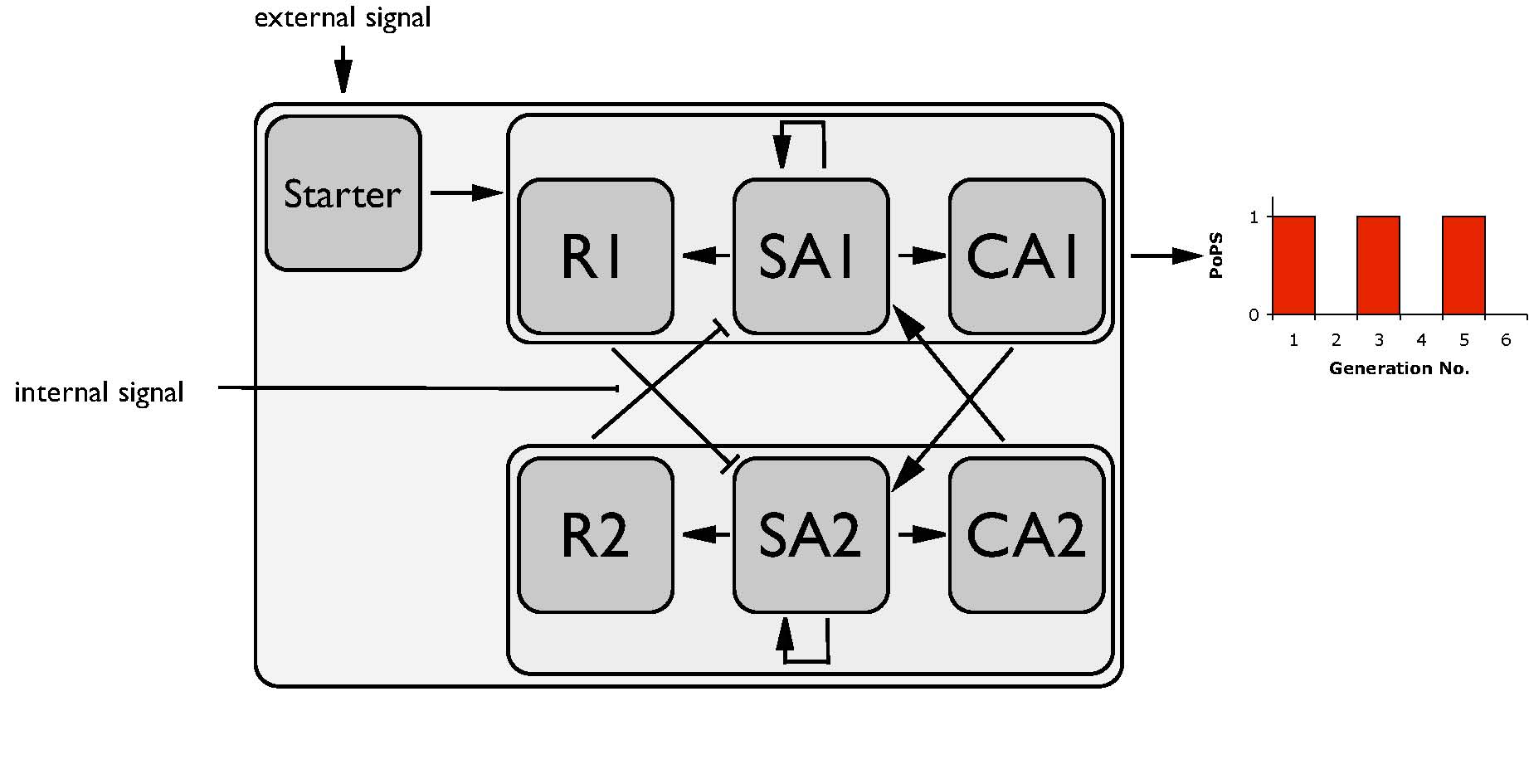
| + | [[Image:ESBS_Device3.jpg|thumb|right|255px|Parts level]] | ||
| + | Both, the odd and the even device are constituted of the same set of three parts, which are in turn assembled parts, constituted from basic Biobrick parts. These are Repressor (R), Self-activator (sA) and Cross-activator (cA). | ||
| + | The sA of each device ensures the expression of all parts of the device. R and cA in turn are both acting on the sA activator of the opposite device. As long as both parts are active (R and cA) the negative effect of R will overlay the positive effect of cA. An internal cell cycle dependent signal will have a negative effect on R, so that cA can induce the sA, thus the expression of the subsystem will be switched.<br> | ||
| + | In order to realize the system, parts have to be assembled from basic Biobrick parts, which fulfil the requirements on each of these three levels. Special attention should be put to the following questions:<br> | ||
| + | *What cell cycle dependent signal can be used to inactivate R?<br> | ||
| + | *How can one ensure that the effect of R is stronger than those of cA?<br> | ||
| + | The modeling has monitored that the system could theoretically work. Please take a closer look on the [https://2008.igem.org/Team:ESBS-Strasbourg/Modeling Modeling] section if you want to know more. | ||
| + | |||
| + | ==Cell cycle dependent Input== | ||
| + | A cell cycle dependent input is required to initiate the switch of our system. Because of the good investigation of the cell cycle in yeast, various inputs which are whether absent or present throughout a time window of the cell cycle are available. This includes cyclins and CDKs (cyclin dependent kinase). When choosing such a cell cycle dependent factor, it is most important that the input is readable for the system in an effective manner. Cell cycle dependent degradation motifs as tags that target a protein to the Anaphase-Promoting Complex (APC) will allow us to authorise the degradation of proteins in a specific time frame. | ||
| - | + | ==Balance of R and cA== | |
| + | The negative effect of the Repressor must overbalance the positive effect of Cross-activator on the self activator of the opposing device. Principally, one could imagine that either a difference in the concentration or in affinity for the promoter could fulfil these requirements. We decided to solve this problem by the use of promoters of different strength. | ||
| + | ==Parts== | ||
| + | As outlined in the "Architecture" section our system is composed of 2 sets having 3 parts each. Following we will state the requirements for these parts and discuss possible choices. | ||
| + | ===Cross Repressor=== | ||
| + | *Requirements | ||
| + | **Negative effect on promoter of opposing construct. | ||
| + | **Degradation in during mitosis, when the APC is active. | ||
| + | *Discussion: | ||
| + | We first of all thought about a pure physical blockade by using truncated transcription factors where only the binding domain will be kept, but soon came across with the TUP1 domain, a protein naturally used in budding yeast to repress the expression of genes. This protein is normally recruited by a set of transcription factors, but we found publications in which a direct fusion of the TUP1 region to a DNA binding domain had a repressive effect. | ||
| + | ===Cross-activator=== | ||
| + | *Requirements: | ||
| + | **Positive Effect on promoter of opposing construct. | ||
| + | **Needs to persist during APC activity, but must be degraded quite fast afterwards. | ||
| + | **Must be dominated by the Cross Repressor in its present | ||
| + | *Discussion: | ||
| + | As we decided to use the same DNA-binding domain for both, the Cross-activator and the Cross-repressor, we needed to construct the Cross-activator in a similar fashion than the Cross-repressor. This is why we decided to use the VP16 activation domain, which is a domain recruiting Transcription Factors and leading finally to the initiation of Transcription, and fuse it to the same DNA-binding domain as the Cross-repressor. | ||
| + | ===Self-activator=== | ||
| + | *Requirements: | ||
| + | **The Self-activator serves to keep the expression of the active construct at a high level, therefore preventing spontaneous switches. Furthermore we wanted that our system is controlled at every point of time. | ||
| + | **Degradation in APC window. | ||
| + | *Discussion: | ||
| + | The Self-activator is exactly the same than the Cross-activator of the other system, with the difference that it is APC tagged. | ||
| - | + | ==Parts Assembly== | |
| + | Knowing the requirements for the individual parts we have to find the optimal assembly of the parts from the basic Biobricks. This process, explained in the [https://2008.igem.org/Team:ESBS-Strasbourg/measurements Measurements] section will give pure system the final shape. | ||
Latest revision as of 00:45, 30 October 2008
System construction
Goal
We intend to construct a cell-cycle dependent toggle switch in eucaryotic cells. This system can then be used as the basis for a binary cell division counter.
Organism
Saccharomyctes cerevisiae is in respect to the cell cycle regulation the best understood model organism. This will give us the possibility to develop our project with high accuracy on a theoretical level. Furthermore, generation times are in the right order to be able to express and also to degrade proteins within one generation time in a predictable manner. There is also the advantage of features like easy transformation and high rates of homologous recombination. In turn, the eucaryotic nature of yeast will allow an easy transfer of the system to more complex eucaryots as for example HeLa cells.
Counting
The mode of binary counting appeared more elegant to us than the continuous way of counting, because the former would need less reporter compounds. So we decided to implement a binary counting system. Thus, this system would allow us to count to a higher number because of the current limitations of reporter constructs. Secondly, the binary counting mechanism would allow us to construct each bit in a modular manner, hence all further bits could be easily attached through minor changes in this standard bit module, the cell cycle dependent toggle switch.
BioBrick Standard
In order to keep the interference between our system and the host metabolism to a minimum we are envisaging the use of yeast parts only where an interaction with the host is unavoidable. There are two unavoidable interfaces which are the dependence on a cell cycle specific signal as input and the use of the host transcriptional and translational machinery to ensure the expression of the system. For all other components we are planning to revert to the vast quantity of procaryotic parts from the Registry. Thus, our project will include various fusion proteins of mixed (eukaryotic, prokaryotic) nature.
By regarding each domain as a part on its own right a rapid construction of components (for example: TFs and Repressors) with different properties would be possible (see also Dhar, PK; 2007). However, it is not be possible to construct fusion proteins by using the BioBrick Standard developed by Knight (Knight, T; 2003) since the 8 base between two parts would lead to a frame shift. Therefore, we envisage the use of the standard developed by Silver (Philips, IE; Silver, PA; 2007) having only six base pairs between two parts. The transformation of existing parts into the Silver standard will therefore be an important part of our project.
In respect to the minimal interference between the system and the host one could even go further by introducing also the whole protein expression machinery from prokaryots. We termed such an approach the e-language.
Architecture
System level
Technical spoken, our goal is to develop a system which, once activated through an external signal, gives us alternately a PoPS output, no output respectively (see figure System level). This system is equal to the first bit of the possible cell division counter, thus would allow us to count to one. According to the idea of synthetic biology, one can use this system by using the adequate PoPS signal without a further understanding of the system's functioning. Thus, the system is available whenever a cell cycle dependent switch is needed.
Device level
The figure on the right is showing our construct architecture on a device level. One can appreciate the functional units of our system. The two devices, odd device and even device, will be alternatively activated in successive generations. By coupling the PoPS outcome of the even device we will obtain the wished expression pattern. Additionally to these two devices, a starter device for the integration of the external start signal and a reporter device (not shown) will be necessary to get constructed.
Parts level
Both, the odd and the even device are constituted of the same set of three parts, which are in turn assembled parts, constituted from basic Biobrick parts. These are Repressor (R), Self-activator (sA) and Cross-activator (cA).
The sA of each device ensures the expression of all parts of the device. R and cA in turn are both acting on the sA activator of the opposite device. As long as both parts are active (R and cA) the negative effect of R will overlay the positive effect of cA. An internal cell cycle dependent signal will have a negative effect on R, so that cA can induce the sA, thus the expression of the subsystem will be switched.
In order to realize the system, parts have to be assembled from basic Biobrick parts, which fulfil the requirements on each of these three levels. Special attention should be put to the following questions:
- What cell cycle dependent signal can be used to inactivate R?
- How can one ensure that the effect of R is stronger than those of cA?
The modeling has monitored that the system could theoretically work. Please take a closer look on the Modeling section if you want to know more.
Cell cycle dependent Input
A cell cycle dependent input is required to initiate the switch of our system. Because of the good investigation of the cell cycle in yeast, various inputs which are whether absent or present throughout a time window of the cell cycle are available. This includes cyclins and CDKs (cyclin dependent kinase). When choosing such a cell cycle dependent factor, it is most important that the input is readable for the system in an effective manner. Cell cycle dependent degradation motifs as tags that target a protein to the Anaphase-Promoting Complex (APC) will allow us to authorise the degradation of proteins in a specific time frame.
Balance of R and cA
The negative effect of the Repressor must overbalance the positive effect of Cross-activator on the self activator of the opposing device. Principally, one could imagine that either a difference in the concentration or in affinity for the promoter could fulfil these requirements. We decided to solve this problem by the use of promoters of different strength.
Parts
As outlined in the "Architecture" section our system is composed of 2 sets having 3 parts each. Following we will state the requirements for these parts and discuss possible choices.
Cross Repressor
- Requirements
- Negative effect on promoter of opposing construct.
- Degradation in during mitosis, when the APC is active.
- Discussion:
We first of all thought about a pure physical blockade by using truncated transcription factors where only the binding domain will be kept, but soon came across with the TUP1 domain, a protein naturally used in budding yeast to repress the expression of genes. This protein is normally recruited by a set of transcription factors, but we found publications in which a direct fusion of the TUP1 region to a DNA binding domain had a repressive effect.
Cross-activator
- Requirements:
- Positive Effect on promoter of opposing construct.
- Needs to persist during APC activity, but must be degraded quite fast afterwards.
- Must be dominated by the Cross Repressor in its present
- Discussion:
As we decided to use the same DNA-binding domain for both, the Cross-activator and the Cross-repressor, we needed to construct the Cross-activator in a similar fashion than the Cross-repressor. This is why we decided to use the VP16 activation domain, which is a domain recruiting Transcription Factors and leading finally to the initiation of Transcription, and fuse it to the same DNA-binding domain as the Cross-repressor.
Self-activator
- Requirements:
- The Self-activator serves to keep the expression of the active construct at a high level, therefore preventing spontaneous switches. Furthermore we wanted that our system is controlled at every point of time.
- Degradation in APC window.
- Discussion:
The Self-activator is exactly the same than the Cross-activator of the other system, with the difference that it is APC tagged.
 "
"