Team:Freiburg/Project
From 2008.igem.org
m |
|||
| Line 17: | Line 17: | ||
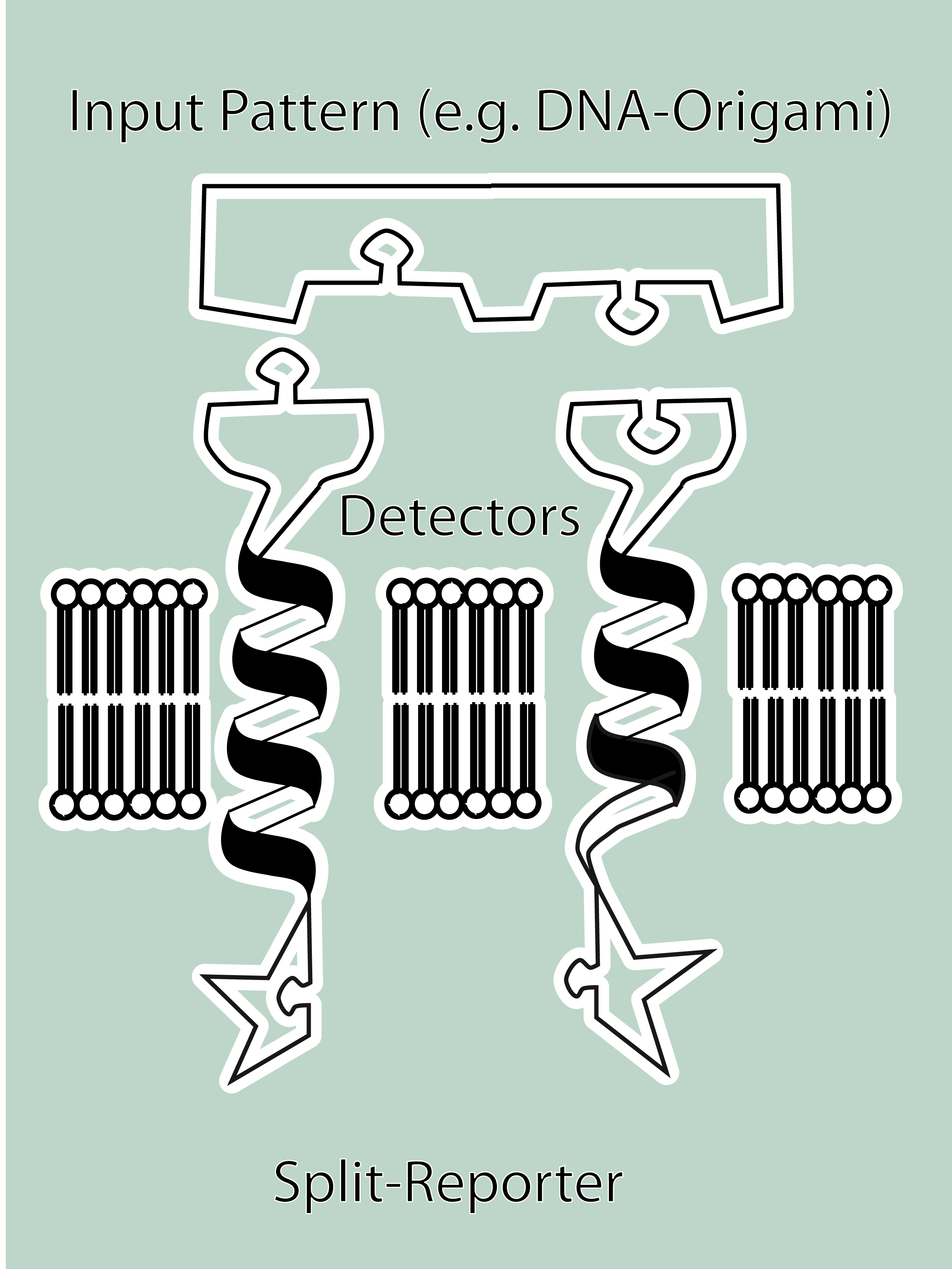
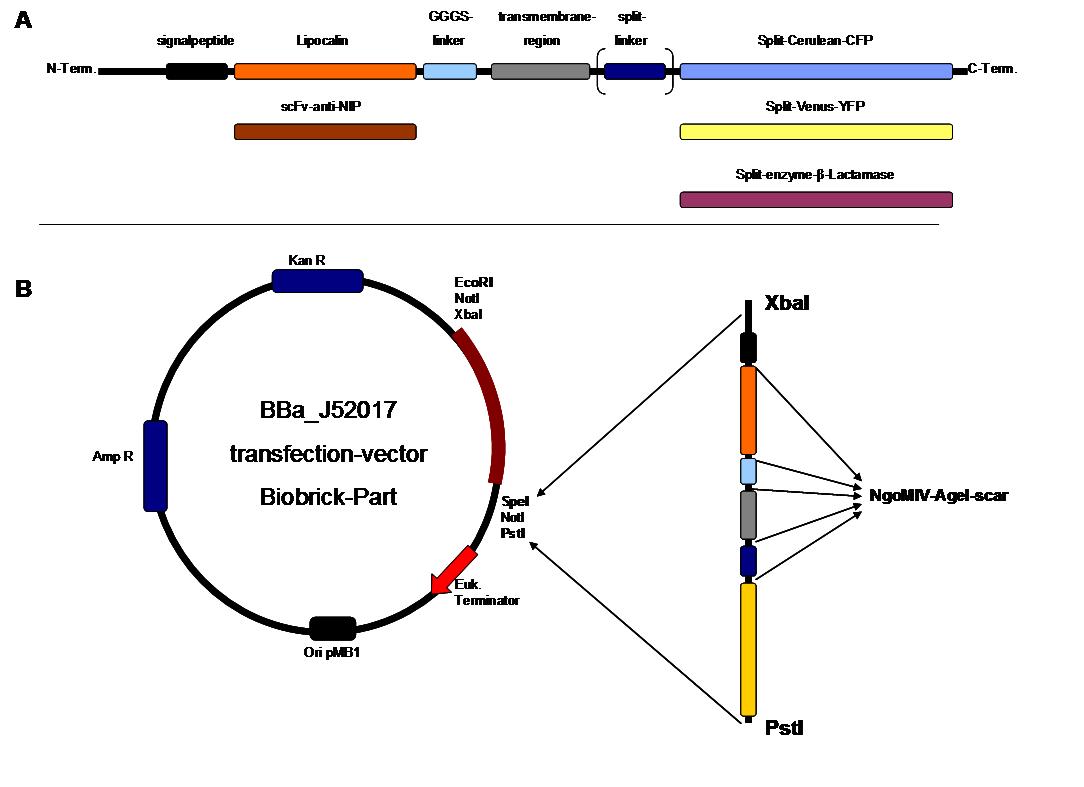
As intracellular reporters we employ split proteins. Here, we used the split lactamase from the iGEM2007 Freiburg project as well as the split fluorescent proteins CFP/cerulean and YFP/venus. To provide steric flexibility and reach of our binding and reporting modules we interspersed linkers between each functional domain.<br> | As intracellular reporters we employ split proteins. Here, we used the split lactamase from the iGEM2007 Freiburg project as well as the split fluorescent proteins CFP/cerulean and YFP/venus. To provide steric flexibility and reach of our binding and reporting modules we interspersed linkers between each functional domain.<br> | ||
| - | All parts submitted feature full BioBrick | + | All parts submitted feature full BioBrick compatibility and in addition allow for the construction of fusion proteins due to extended Biobrick 3.0 (Freiburg) standard. Cloning was done in <i>E. coli</i>. However, we test our receptors in human or mouse cells/chassis. Thus, to direct our constructs to the cell surface we attached a eukaryotic signal peptide taken from hEGF-R to the N-terminus of each synthetic receptor. In addition to our designed synthetic receptors, we initially also employed an existing T-cell line.<br> |
| - | To reach our goal within the short given time frame we started several subprojects in parallel. Our subprojects listed here are defined along these projects. Besides designing and cloning parts we spent quite some time on establishing and analyzing the biological experiments. Initially we tested interfacing of DNA origami with cells and the spatial control via DNA origami using | + | To reach our goal within the short given time frame we started several subprojects in parallel. Our subprojects listed here are defined along these projects. Besides designing and cloning parts we spent quite some time on establishing and analyzing the biological experiments. Initially we tested interfacing of DNA origami with cells and the spatial control via DNA origami using the existing T-cell line expressing the scFv fused to a T-cell receptor. For T-cell receptor experiments we used Ca2+ signaling a read out. Synthetic receptor activation was analyzed using the formation of fluorescent proteins or turnover of a fluorescent lactamase substrate. In our experiments we addressed the following questions:<br> |
- Can we improve DNA origami assembly yield by varying the staple oligonucleotide to scaffold DNA ratio?<br> | - Can we improve DNA origami assembly yield by varying the staple oligonucleotide to scaffold DNA ratio?<br> | ||
| Line 49: | Line 49: | ||
<td> | <td> | ||
<h4>[[Team:Freiburg/Modeling|Modeling]]</h4> | <h4>[[Team:Freiburg/Modeling|Modeling]]</h4> | ||
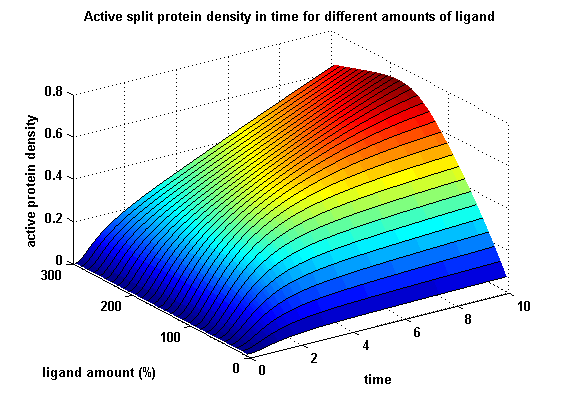
| - | + | Dimerization of extracellular receptor domains is an important necessity for the functionality of our Modular Synthetic Receptor System. Presenting the system a stimulus in form of spatially arranged ligands (nitro-iodo-phenol or fluorescein molecules) results in dimerization and thus the corresponding intracellular parts such as the split lactamase halves or split fluorescent proteins complement to measureable output. To analyze functionality due to dimerization, two receptor dimerization models (one T cell receptor model and one general receptor model) are introduced and discussed and a proper model for the Modular Synthetic Receptor System is constructed. It consists of 10 reaction kinetic equations, 9 ordinary differential equations including 25 variable parameters. Matlab m-files are embedded for further inspection. | |
</td> | </td> | ||
<td> | <td> | ||
| Line 64: | Line 64: | ||
<td> | <td> | ||
<h4>[[Team:Freiburg/3D-Modeling|3D-Modeling]]</h4> | <h4>[[Team:Freiburg/3D-Modeling|3D-Modeling]]</h4> | ||
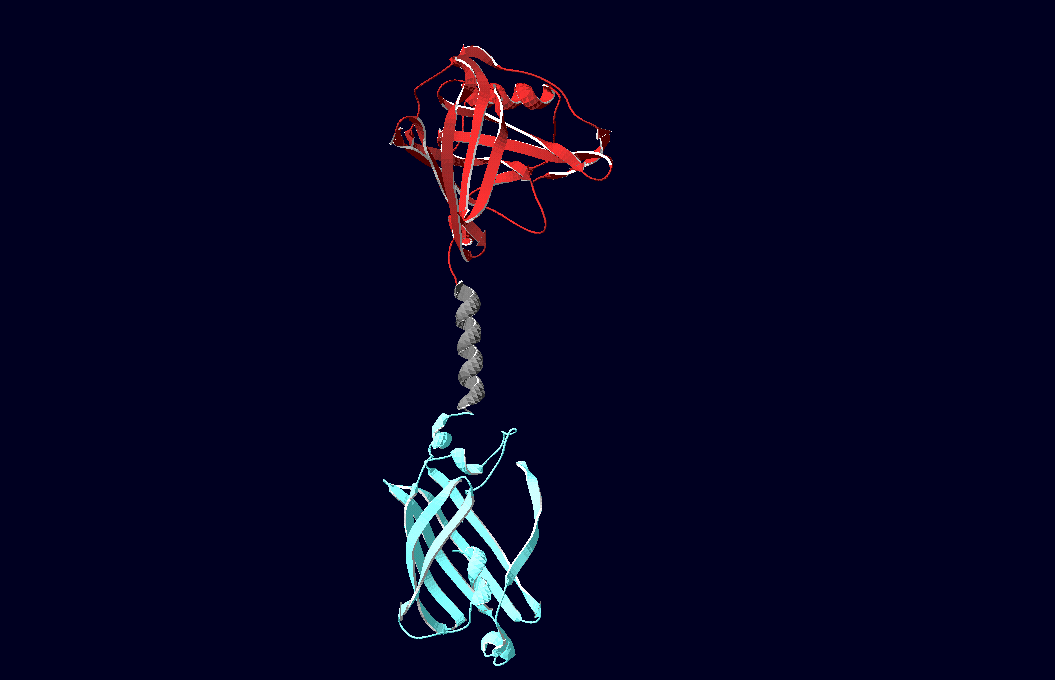
| - | We have created various three-dimensional models of our constructs using Pymol and SwissPdbViewer. | + | We have created various three-dimensional models of our constructs using Pymol and SwissPdbViewer. Pdb-files of the huge DNA-Origami molecule were created as adequate as possible using Nano-Engineer version 1.0 and then edited in Pymol. These Models were used to plan the spatial arrangement and, thus, input-pattern of antigens at a nanometer scale.<br><br><br> |
| - | Pdb-files of the huge DNA-Origami molecule were created as adequate as possible using Nano-Engineer version 1.0 and then edited in Pymol. These Models were used to plan the spatial arrangement and, thus, input-pattern of antigens at a nanometer scale.<br><br><br> | + | |
</td> | </td> | ||
</tr> | </tr> | ||
| Line 74: | Line 73: | ||
<td> | <td> | ||
<h4>[[DNA-Origami|DNA-Origami]]</h4> | <h4>[[DNA-Origami|DNA-Origami]]</h4> | ||
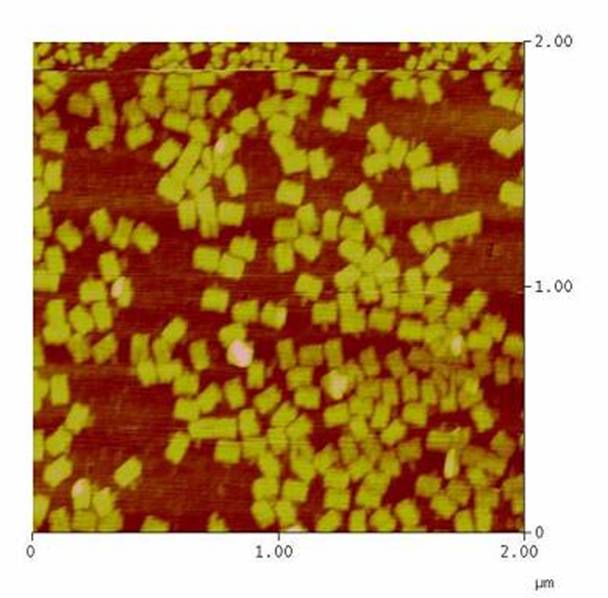
| - | The creation of DNA-Origami was one of the first sub-projects we engaged in. Structural planing of the input pattern on the Origami-surface and order of the modified oligo-nucleotides were the first issues, increasing of the Origami-yield and measurement of the molecule´s stability in cell culture media the | + | The creation of DNA-Origami was one of the first sub-projects we engaged in. Structural planing of the input pattern on the Origami-surface and order of the modified oligo-nucleotides were the first issues, increasing of the Origami-yield and measurement of the molecule´s stability in cell culture media the later ones.<br> |
</td> | </td> | ||
<td> | <td> | ||
| Line 99: | Line 98: | ||
<td> | <td> | ||
<h4>[[Team:Freiburg_Transfection and Synthetic Receptor|Transfection and Synthetic Receptor Activation]]</h4> | <h4>[[Team:Freiburg_Transfection and Synthetic Receptor|Transfection and Synthetic Receptor Activation]]</h4> | ||
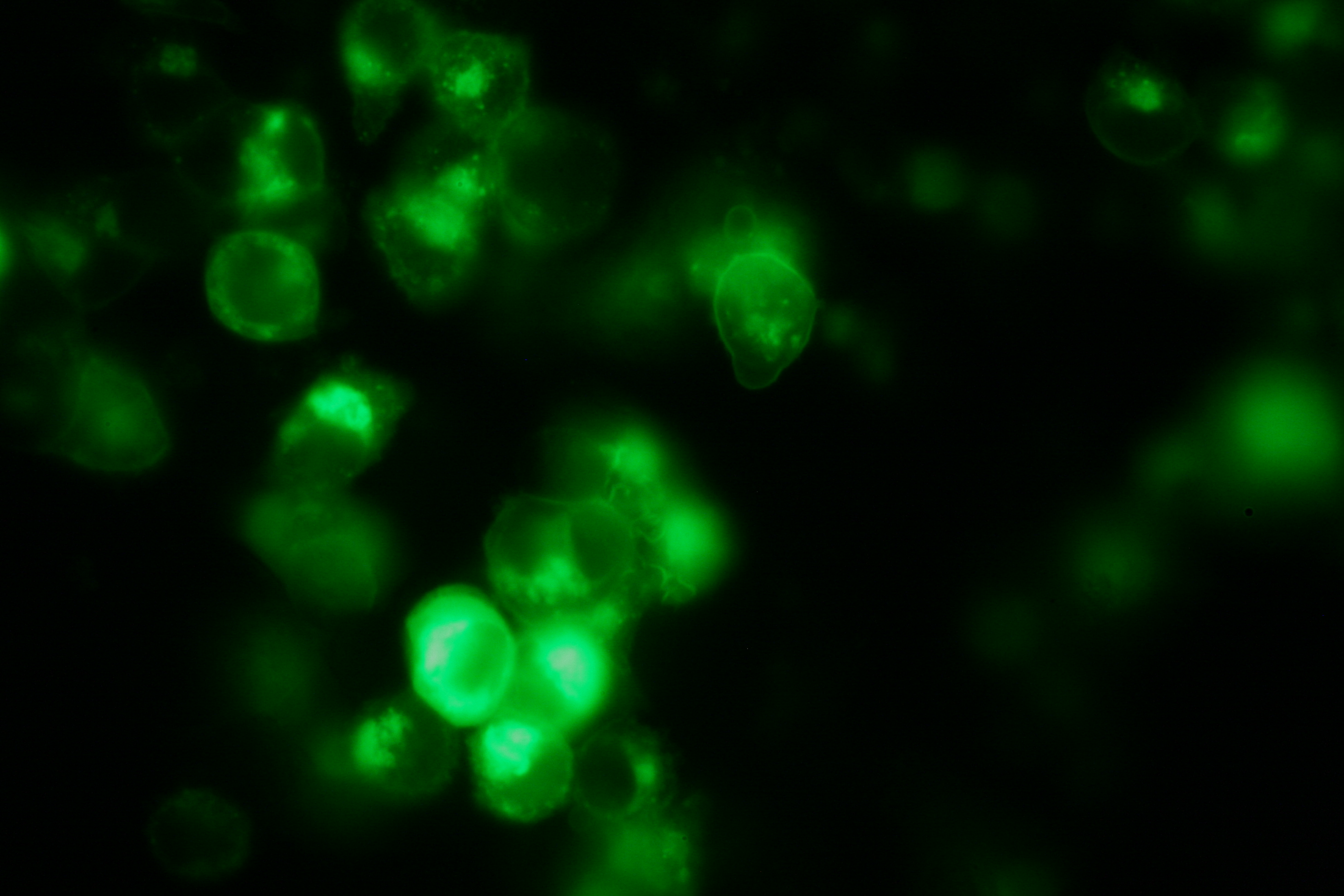
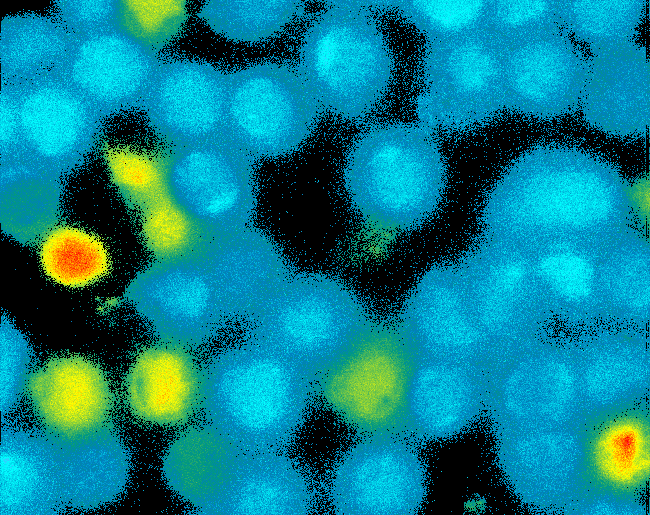
| - | Transfection of 293T-cells has | + | Transfection of 293T-cells has been shown to work for most of our fusion proteins; so far, we could even show that at least our constructs with lipocalin FluA as extracellular domaine are integrated into the membrane.<br> |
| - | All transfections were carried out with part Bba_K157040, one of our composite parts consisting of the transfection vector Bba_J52017 and a CMV-promotor. Due to the limited time range we were not able to | + | All transfections were carried out with part Bba_K157040, one of our composite parts consisting of the transfection vector Bba_J52017 and a CMV-promotor. Due to the limited time range we were not yet able to demonstrate activation of the various possible "receptor"-pairs, but we are hoping to receive a positive result soon.<br> |
</td> | </td> | ||
<td> | <td> | ||
Revision as of 01:07, 30 October 2008
|
Project Report |
_project report
SummaryModular Synthetic Receptor System In the following we provide a summary of our project and display the highlights of our achievements. For a more detailed view please see the reports of each subproject. The general goal of the Freiburg 2008 team is to establish a synthetic transmembrane receptor system comprising the extracellular input devices, the extracellular receiver domain, the transducer to the cytosol and the reporter/executor in the cytosol. To facilitate binding, we used a single-chain Fv antibody fragment and a designed anticalin. Both bind haptens (nitro-jodo-phenol and fluorescein) which can be conjugated to specific oligonucleotides of the DNA-Origami or scaffold proteins. Transduction to the cytosol is mediated by a single transmembrane helix. As intracellular reporters we employ split proteins. Here, we used the split lactamase from the iGEM2007 Freiburg project as well as the split fluorescent proteins CFP/cerulean and YFP/venus. To provide steric flexibility and reach of our binding and reporting modules we interspersed linkers between each functional domain. All parts submitted feature full BioBrick compatibility and in addition allow for the construction of fusion proteins due to extended Biobrick 3.0 (Freiburg) standard. Cloning was done in E. coli. However, we test our receptors in human or mouse cells/chassis. Thus, to direct our constructs to the cell surface we attached a eukaryotic signal peptide taken from hEGF-R to the N-terminus of each synthetic receptor. In addition to our designed synthetic receptors, we initially also employed an existing T-cell line. To reach our goal within the short given time frame we started several subprojects in parallel. Our subprojects listed here are defined along these projects. Besides designing and cloning parts we spent quite some time on establishing and analyzing the biological experiments. Initially we tested interfacing of DNA origami with cells and the spatial control via DNA origami using the existing T-cell line expressing the scFv fused to a T-cell receptor. For T-cell receptor experiments we used Ca2+ signaling a read out. Synthetic receptor activation was analyzed using the formation of fluorescent proteins or turnover of a fluorescent lactamase substrate. In our experiments we addressed the following questions: - Can we improve DNA origami assembly yield by varying the staple oligonucleotide to scaffold DNA ratio? In our modeling analyses we constructed various sets of differential equations for our synthetic receptors and predicted split protein activation behaviour. The labs of Kristian Müller and Katja Arndt provided all basic technology and support to get started. For advanced analyses several labs of the University of Freiburg and the Max-Planck-Institute of Immunobiology granted access to their instrumentation (e.g. atomic force micoscopy, confocal microscopy, FACS).
SubprojectsModeling Highlights
|
 "
"