Team:Freiburg/Project
From 2008.igem.org
m |
m |
||
| (2 intermediate revisions not shown) | |||
| Line 21: | Line 21: | ||
To reach our goal within the short given time frame we started several subprojects in parallel. Our subprojects listed here are defined along these projects. Besides designing and cloning parts we spent quite some time on establishing and analyzing the biological experiments. Initially we tested interfacing of DNA origami with cells and the spatial control via DNA origami using the existing T-cell line expressing the scFv fused to a T-cell receptor. For T-cell receptor experiments we used Ca<sup>2+</sup> signaling as read out. Synthetic receptor activation was analyzed using the formation of fluorescent proteins or turnover of a fluorescent lactamase substrate. In our experiments we addressed the following questions:<br> | To reach our goal within the short given time frame we started several subprojects in parallel. Our subprojects listed here are defined along these projects. Besides designing and cloning parts we spent quite some time on establishing and analyzing the biological experiments. Initially we tested interfacing of DNA origami with cells and the spatial control via DNA origami using the existing T-cell line expressing the scFv fused to a T-cell receptor. For T-cell receptor experiments we used Ca<sup>2+</sup> signaling as read out. Synthetic receptor activation was analyzed using the formation of fluorescent proteins or turnover of a fluorescent lactamase substrate. In our experiments we addressed the following questions:<br> | ||
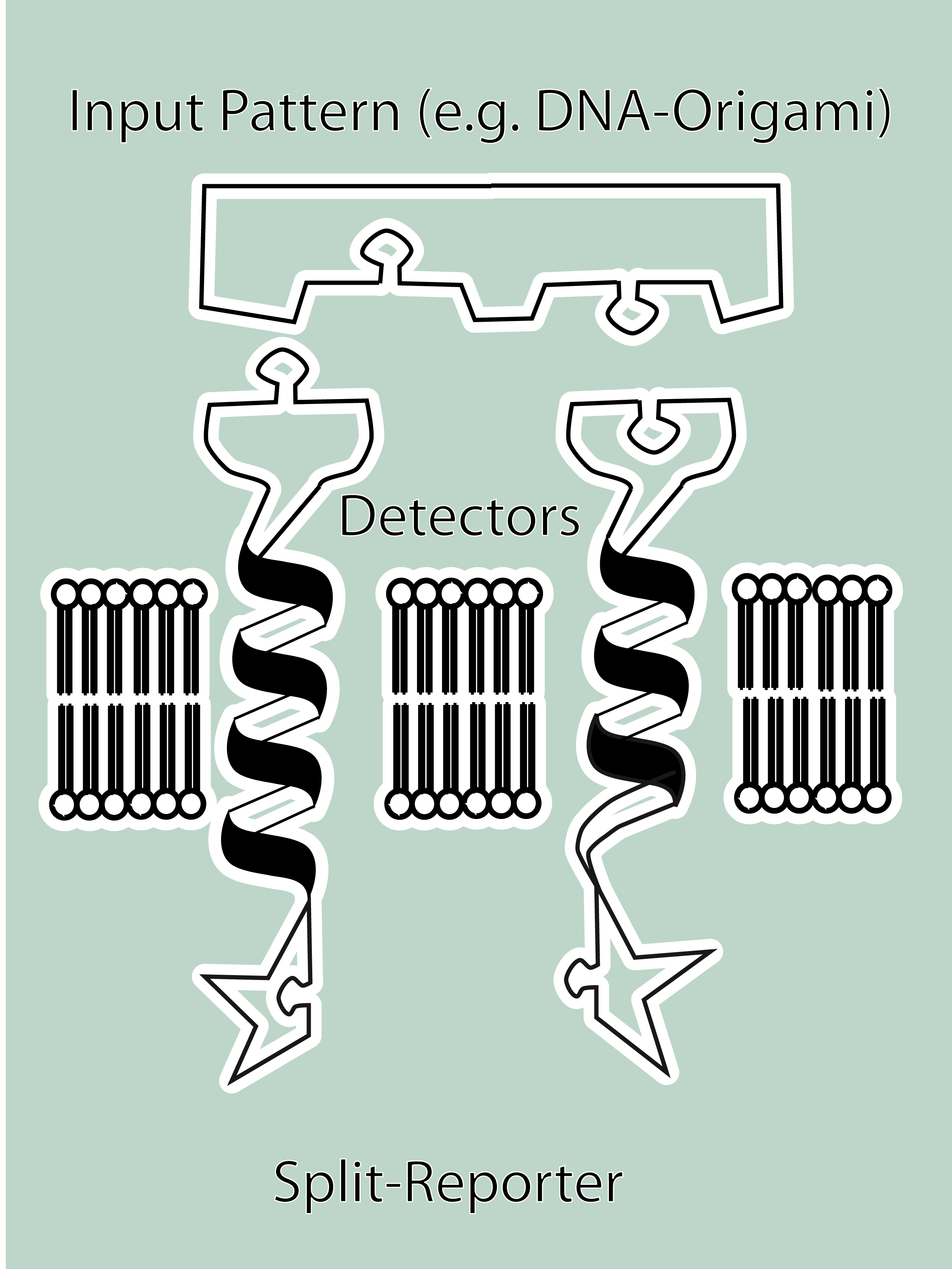
| - | + | * Can we design DNA origami to induce receptor multimerization?<br> | |
| - | + | * Can we improve DNA origami assembly yield by varying the staple oligonucleotide to scaffold DNA ratio?<br> | |
| - | + | * Can we find buffer conditions, which mediate cell viability and origami stability?<br> | |
| - | + | * Can we downsize a Ca<sup>2+</sup> release assay to fit the sample sizes of our precious DNA origami?<br> | |
| - | + | * Do our constructs express and then localize in the membrane of eukaryotic cells?<br> | |
| - | + | * Do DNA origamis bind to cells?<br> | |
| - | + | * Can we detect synthetic receptor activation?<br> | |
In short, we successfully addressed the first six questions. Experiments to demonstrate synthetic receptor activation are ongoing. | In short, we successfully addressed the first six questions. Experiments to demonstrate synthetic receptor activation are ongoing. | ||
| Line 114: | Line 114: | ||
<tr> | <tr> | ||
<td> | <td> | ||
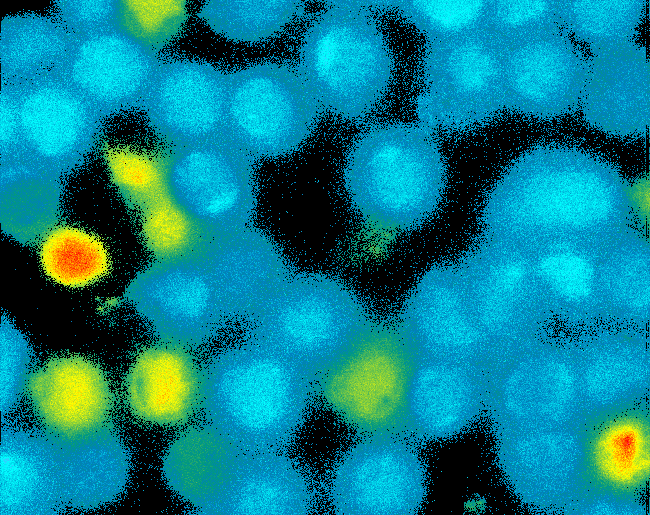
| - | [[Image:TeamFreiburg2008_Pervanadate1.png|thumb|left|200px|Ca-influx measurement]] | + | [[Image:TeamFreiburg2008_Pervanadate1.png|thumb|left|200px|Ca<sup>2+</sup>-influx measurement]] |
</td> | </td> | ||
<td> | <td> | ||
| Line 138: | Line 138: | ||
*Ingo P. Korndörfer, Gerald Beste and A. Skerra: “Crystallographic Analysis of an “Anticalin” With Tailored Specificity for Fluorescein Reveals High Structural Plasticity of the Lipocalin Loop Region”, PROTEINS: Structure, Function, and Bioinformatics 53:121–129 (2003)<br> | *Ingo P. Korndörfer, Gerald Beste and A. Skerra: “Crystallographic Analysis of an “Anticalin” With Tailored Specificity for Fluorescein Reveals High Structural Plasticity of the Lipocalin Loop Region”, PROTEINS: Structure, Function, and Bioinformatics 53:121–129 (2003)<br> | ||
'''DNA-Origami:'''<br> | '''DNA-Origami:'''<br> | ||
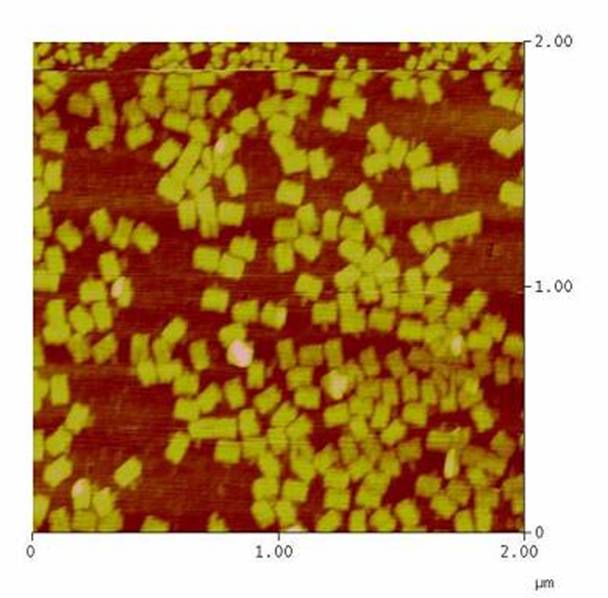
| - | Paul W. K. Rothemund: "Folding DNA to create nanoscale shapes and patterns", Nature 440, 297-302 (16 March 2006)<br> | + | *Paul W. K. Rothemund: "Folding DNA to create nanoscale shapes and patterns", Nature 440, 297-302 (16 March 2006)<br> |
'''Antibody B1-8:'''<br> | '''Antibody B1-8:'''<br> | ||
*Ana Cumano and Klaus Rajewski: “Clonal recruitment and somatic mutation in the generation of immunological memory to the hapten NP”, The EMBO Journal vol. 5 no.10 pp. 2459-2468, 1986<br> | *Ana Cumano and Klaus Rajewski: “Clonal recruitment and somatic mutation in the generation of immunological memory to the hapten NP”, The EMBO Journal vol. 5 no.10 pp. 2459-2468, 1986<br> | ||
Latest revision as of 08:24, 30 October 2008
|
Project Report |
_project report
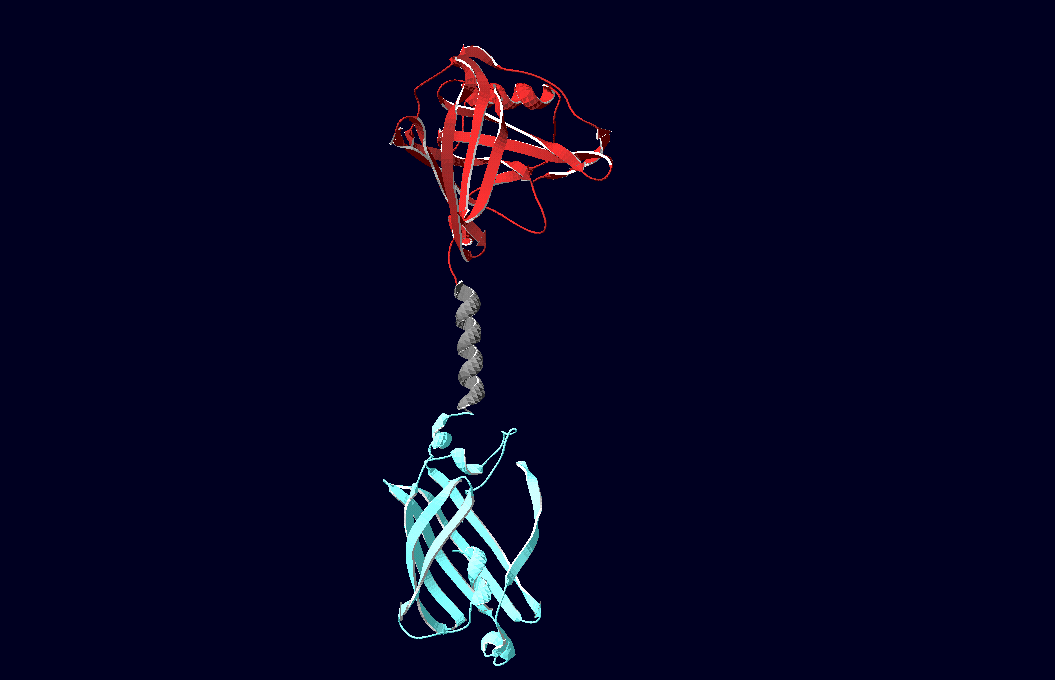
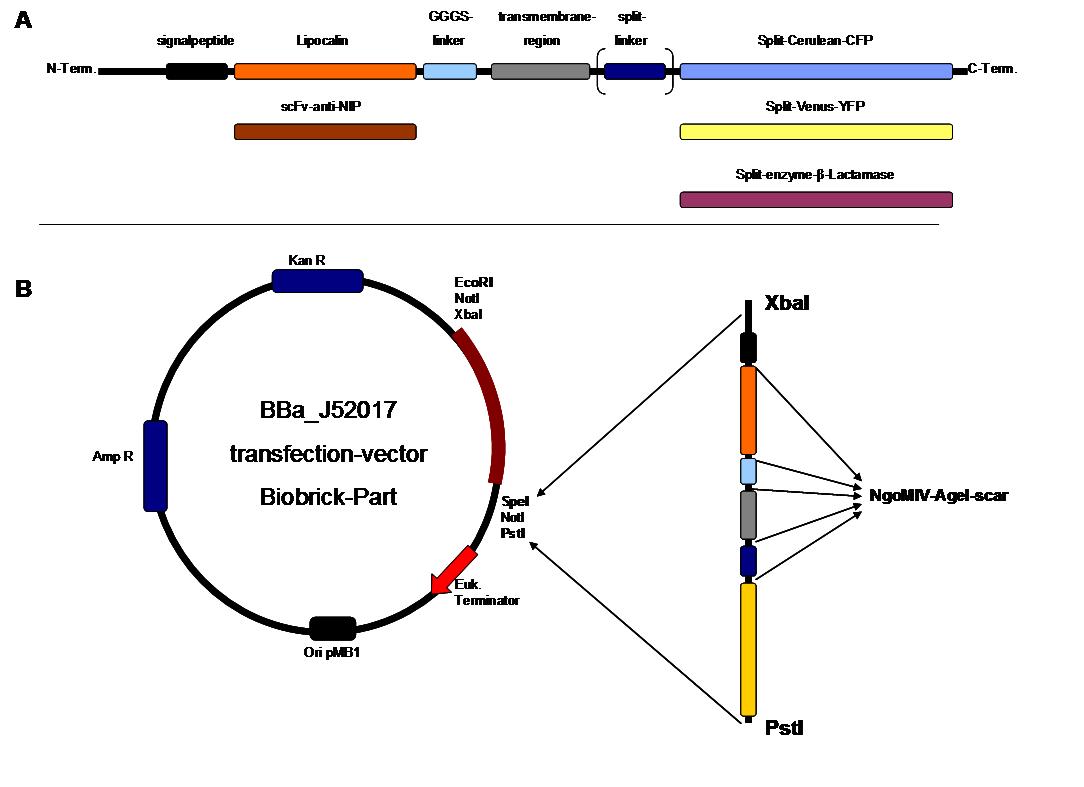
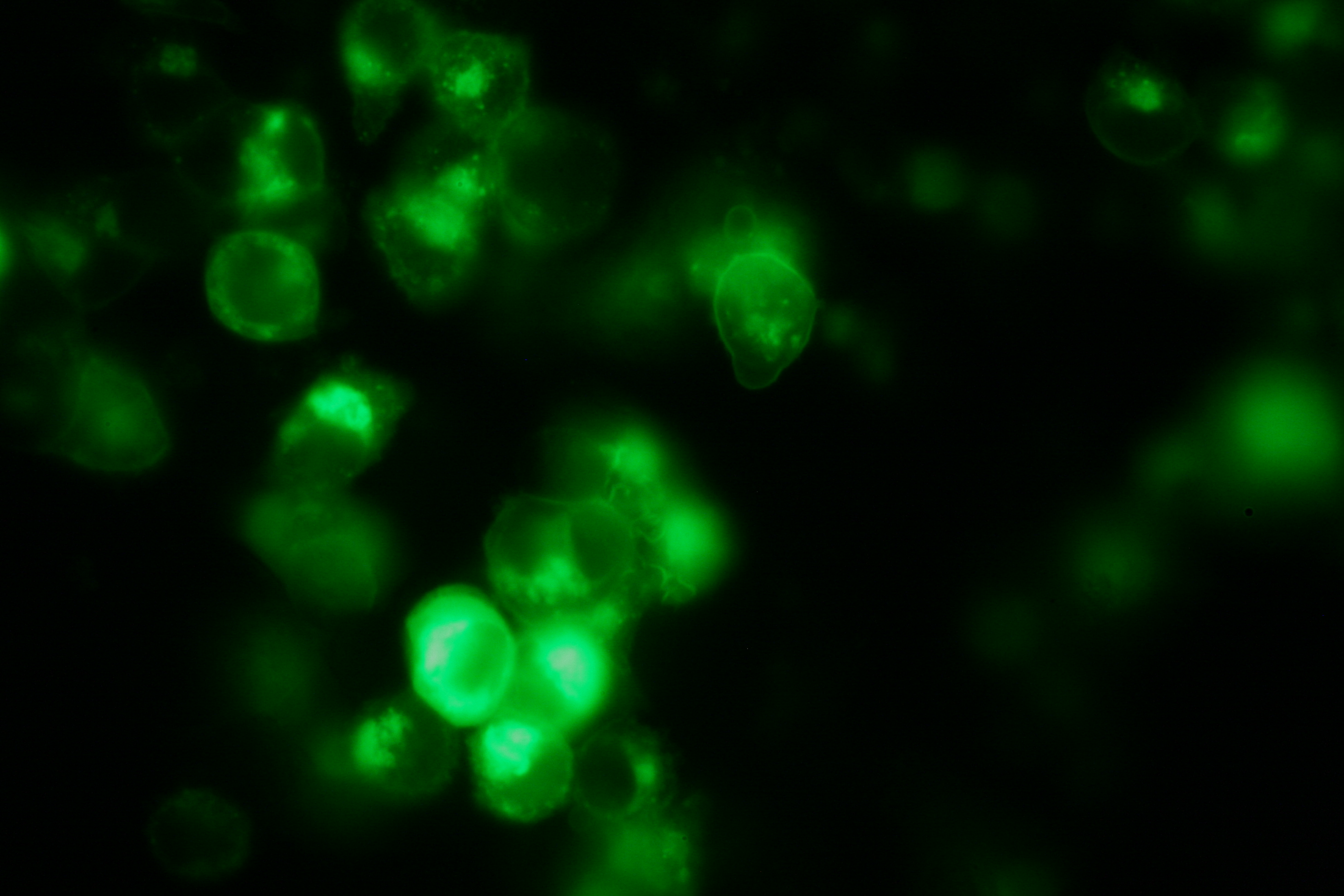
SummaryModular Synthetic Receptor System In the following we provide a summary of our project and display the highlights of our achievements. For a more detailed view please see the reports of each subproject. The general goal of the Freiburg 2008 team is to establish a synthetic transmembrane receptor system comprising the extracellular input devices, the extracellular receiver domain, the transducer to the cytosol and the reporter/executor in the cytosol. To facilitate binding, we used a single-chain Fv antibody fragment and a designed anticalin. Both bind haptens (nitro-jodo-phenol and fluorescein) which can be conjugated to specific oligonucleotides of the DNA origami or scaffold proteins. Transduction to the cytosol is mediated by a single transmembrane helix taken either from the EGF-receptor or B-cell-receptor. As intracellular reporters we employ split proteins. Here, we used the split lactamase from the iGEM2007 Freiburg project as well as the split fluorescent proteins CFP/cerulean and YFP/venus. To provide steric flexibility and reach of our binding and reporting modules we interspersed linkers between each functional domain. All parts submitted feature full BioBrick compatibility and in addition allow for the construction of fusion proteins due to extended Biobrick 3.0 (Freiburg) standard. Cloning was done in E. coli. However, we test our receptors in human or mouse cells/chassis. Thus, to direct our constructs to the cell surface we attached a eukaryotic signal peptide taken from hEGF-R to the N-terminus of each synthetic receptor. In addition to our designed synthetic receptors, we initially also employed an existing T-cell line. To reach our goal within the short given time frame we started several subprojects in parallel. Our subprojects listed here are defined along these projects. Besides designing and cloning parts we spent quite some time on establishing and analyzing the biological experiments. Initially we tested interfacing of DNA origami with cells and the spatial control via DNA origami using the existing T-cell line expressing the scFv fused to a T-cell receptor. For T-cell receptor experiments we used Ca2+ signaling as read out. Synthetic receptor activation was analyzed using the formation of fluorescent proteins or turnover of a fluorescent lactamase substrate. In our experiments we addressed the following questions:
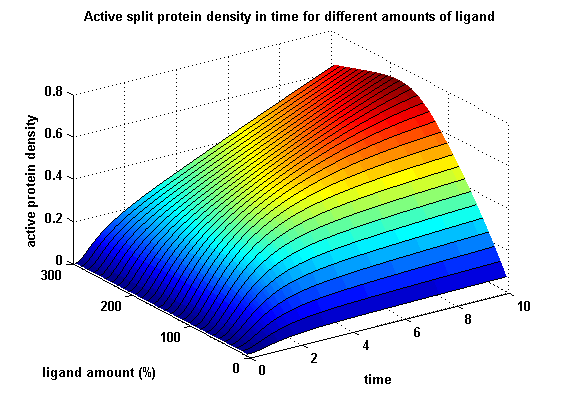
In short, we successfully addressed the first six questions. Experiments to demonstrate synthetic receptor activation are ongoing. For our modeling analyses we constructed various sets of differential equations describing our synthetic receptors and predicted split protein activation behaviour. The labs of Kristian Müller and Katja Arndt provided all basic technology and support to get started. For advanced analyses several labs of the University of Freiburg and the Max-Planck-Institute of Immunobiology granted access to their instrumentation (e.g. atomic force micoscopy, confocal microscopy, FACS).
Subprojects
Highlights
|
 "
"