Team:Waterloo/Project
From 2008.igem.org
| (44 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
<!--- The Mission, Experiments ---> | <!--- The Mission, Experiments ---> | ||
| - | {| style="color:#1b2c8a;background-color:# | + | {| style="color:#1b2c8a;background-color:#FBCC30;" cellpadding="3" cellspacing="1" border="1" bordercolor="#fff" width="62%" align="center" |
!align="center"|[[Team:Waterloo|Home]] | !align="center"|[[Team:Waterloo|Home]] | ||
!align="center"|[[Team:Waterloo/Team|The Team]] | !align="center"|[[Team:Waterloo/Team|The Team]] | ||
| Line 8: | Line 8: | ||
!align="center"|[[Team:Waterloo/Modeling|Modeling]] | !align="center"|[[Team:Waterloo/Modeling|Modeling]] | ||
!align="center"|[[Team:Waterloo/Notebook|Notebook]] | !align="center"|[[Team:Waterloo/Notebook|Notebook]] | ||
| + | !align="center"|[[Team:Waterloo/Sponsors|Sponsors]] | ||
|} | |} | ||
| + | <br> | ||
| - | == ''' | + | == '''Project Aim and Rationale''' == |
| - | + | There are many useful bioproducts that can be efficiently expressed <i>in vivo</i> but are either cost-prohibitive or impossible to synthesiize and purify <i>in vitro</i>. To address this need for a means of manufacturing such bioproducts, a great deal of work had been done to study [http://openwetware.org/wiki/Minicells minicells], genome-free sacks of cytoplasm that bud off from certain mutant bacterial strains, and from which plasmid-driven expression has previously shown to be possible. Minicells showed substantial promise for possible therapeutic applications since they could generate useful products without replicating themselves. However, for <i>in situ</i> delivery of their products, they must be filtered from the live bacteria from which they were released. | |
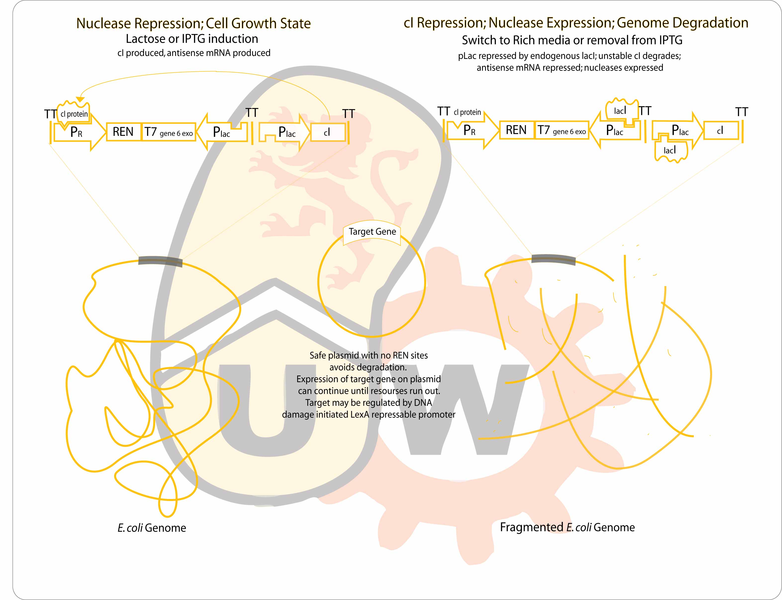
| - | The primary application of this would be an in situ compound production and delivery system for agricultural | + | Transient plasmid-driven expression in minicells has been shown to last for several hours, and though this "lifetime" can be improved by use of a nutrient bath, it is limited by cytosolic resources. Once these resources are exhausted, the minicell becomes inactive and degrades. |
| + | |||
| + | Our project improves on the two issues described above. A bacterial cell already containing a plasmid encoding bioproduct synthesis genes, will self-destruct by degrading its own genome and transiently produce the bioproduct until cell resources have been exhausted. Having the original bacteria serve as the factory means that not only is there no live parental budding strain to purify from the culture, but also more cell resources are available to express larger quantities of bioproduct. | ||
| + | |||
| + | Genome degradation is achieved using the combined activity of a restriction endonuclease to fragment the genome and an exonuclease to hasten degradation. The gene for the protein of interest will be located on a plasmid lacking recognition sites for the endonuclease, allowing it to remain intact after genomic degradation. The plasmid genes will be expressed using the remaining cell resources until they expire. The primary application of this design would be an in situ compound production and delivery system for agricultural, industrial or therapeutic use. | ||
| + | |||
| + | == Project Design == | ||
[[Image:2008Design.png]] | [[Image:2008Design.png]] | ||
| + | === Production and Regulation of Nuclease Genes === | ||
| + | |||
| + | *Endo- and exonucleases will be cloned between the lambda pR promoter and the Plac promoter. The lambda promoter will drive the production of the forward transcript for translation of the genes (which are toxic), while pLac will drive production of antisense transcripts to maximize repression of the two nucleases during uninduced conditions. Cultures will initially be grown on media containing IPTG. Expression of the unstable cI repressor (C0051) will be driven by another instance of a lacI repressable promoter. Since nuclease production is usually repressed, endogenous lacI production should be sufficient. Any antisense transcripts from the leaky Plac will not be able to compete with the strong lambda promoter during expression of the nucleases. | ||
| + | |||
| + | ==Construction Strategy== | ||
| + | |||
| + | ===Biobrick Part Number Legend=== | ||
| + | |||
| + | *[TT] BBa_B0015 | ||
| + | *[Pr] BBa_R0051 | ||
| + | *[Plac] BBa_R0011 | ||
| + | *[CI] BBa_C0051 | ||
| + | *[T7gene6] BBa_K093001 | ||
| + | *[PmeI] BBa_K093002 | ||
| + | |||
| + | ===Steps in Assembly=== | ||
| + | *T7 gene 6 exonuclease will be amplified using PCR with primer-adapters for the BioBrick Prefix and Suffix. | ||
| + | *''Pseudomonas mendocina'' PmeI endonuclease will be synthesized with its ORF flanked by BioBrick Prefix and Suffix. | ||
| + | |||
| + | *A <b>nuclease construct</b> will be constructed using crossover PCR of the nuclease gene. | ||
| + | |||
| + | [[Image:Sequence_(1).jpg]] | ||
| + | |||
| + | *A <b> convergent promoter system</b> designed to flank the nuclease genes are denoted below by Construct 1 and Construct 2. Construct 2 will be added downstream of the nuclease constructs such that no sense transcript and protein will be made at this stage. Construct 1 will be added upstream in the next round of assembly. This will ensure repression of the system in the presence of CI repressor and IPTG. | ||
| + | |||
| + | [[Image:Sequence_(2).jpg]] Construct 1<br> | ||
| + | [[Image:Sequence_(3).jpg]] Construct 2 | ||
| + | |||
| + | *The <b>addition of Construct 2</b> is a non toxic intermediate step. | ||
| + | |||
| + | [[Image:1Sequence_(4).jpg]] | ||
| + | |||
| + | *The <b>addition of Construct 1</b> ensures for proper repression in the presence of IPTG and the CI repressor. | ||
| + | |||
| + | [[Image:1Sequence_(5).jpg]] | ||
| + | |||
| + | *The already assembled CI repressor under the control of Plac must be <b>cotransformed</b> and <b>plated on IPTG</b> | ||
| + | |||
| + | *The <b>final independent construct</b>: | ||
| + | |||
| + | [[Image:1Sequence_(6).jpg]] | ||
| + | |||
| + | *These tests will determine: | ||
| + | **The length of time needed for the destruction of the genome by nuclease expression | ||
| + | **The optimal integration site based on the ideal length of time for nuclease gene production. | ||
| + | **Where the REN sites are on the genome. | ||
| + | **The rate of degradation by the exonuclease. | ||
| + | |||
| + | Note: | ||
| + | *Ligation scars are not shown. | ||
| + | *Ribosome binding sites are present for open reading frames (T7gene6, PmeI, CI) upstream in the forward orientation. | ||
| + | *[forward arrow] denotes forward orientation | ||
| + | *[reverse arrow] denotes reverse position. | ||
| + | |||
| + | ==Implementation== | ||
| + | ===Preliminary Assay of Genomic Degradation=== | ||
| + | *Chassis will be transformed with the preliminary construct. | ||
| + | *Uninduced cell growth will be assayed to determine if the repression is strong enough to allow for the uninhibited growth of uninduced cells. | ||
| + | *Cells will be induced for different periods of time. The induction will be terminated by washing to eliminate IPTG. Assay for sterility of culture growth test with positive and negative controls (uninduced transformants and no inoculation, respectively). Genome destruction and plasmid safeness will be checked by genomic and plasmid preps of the induced culture. [(looking for smear and nice band?)] | ||
| + | |||
| + | ===Integration of Nuclease Genes=== | ||
| + | *The genome degradation construct will be integrated into the genome via a proposed standard integration vector. | ||
| + | *The vector will contain a BioBrick cloning site and unique double digest sites flanking the BioBrick cloning site and suitable elements for selection (Sac B and antibiotic resistance). The unique double digest sites will allow the user to customize the regions of homology for a double crossover homologous recombination. | ||
| + | |||
| + | *The integration vector will be a vector with the oriR6k origin of replication that only replicates in ''E. coli'' lambda pir lysogen. For integration, the plasmid will be transformed into a strain of ''E. coli'' that is not a lambda pir lysogen by selecting for recombination using the antibiotic respective to the vector. The vector will be a suicide vector and therefore in any non lambda pir strain, a single recombination will be selected. The cells will then be removed from selective pressure to allow for the next recombination to occur. The double recombinants will be selected using sucrose, since the vector will also contain a ''sacB'' gene, which confers sucrose sensitivity. The putative double recombinants must then be screened for the respective antibiotic sensitivity. | ||
| - | == | + | ===Safe Target Plasmid=== |
| - | * | + | *In order to ensure expression of a desired gene post degradation of the genome, the plasmid must be free of recognition sites for the restriction endonuclease PmeI and therefore be unable to be influenced by the exonuclease. Red fluorescent protein is used as a reporter gene. |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| + | *The reporter will be put under the control of the LexA repressible promoter. The genome degradation method produces single stranded DNA due to the presence of the 5’-3’ [(check gene 6 exo activity direction to confirm)] exonuclease. When there is single stranded DNA in the cell, RecA is upregulated as part of the SOS response, and RecA cleaves lexA. Therefore when the genome degradation is induced, the expression of the gene of interest will be subsequently induced. For this reason, the final construct must be integrated into a RecA + strain (TG1 is Rec+ and lacIq, and will work completely since lacI is required for the induction of the genome degradation operon). | ||
| + | *The plasmid safeness will be assayed when exposed to the nucleases. From that, reporter production can be evaluated, and the induction time relative to the time of degradation can be optimized. | ||
| - | == | + | ==Preliminary Testing of Promoter Reporter Constructs== |
| + | A preliminary experiment was conducted to test the expression of the pRecA promoter with LexA binding sites (K093000), the constitutive promoter pconst (J23118) and PocI, the lambda Pr promoter containing the operator for lambda cI repressor (K093010). Visual observations of liquid cultures show that expression of RFP under the control of the PocI promoter in absence of the repressor is high and similar to expression under control of the constitutive promoter. These cultures were all extremely pink, and also form pink colonies on solid media. The uninduced pRecA culture shows very low levels of expression and light pink colonies. | ||
| + | Cultures were inoculated in triplicate. Saturated overnight cultures were inoculated from a single colony. A second set of cultures were inoculated and grown to an OD600 of approximately 0.8. For pRecA, 3 conditions were set and conducted at both cell concentrations; uninduced culture, culture exposed to 125V of UV culture exposed to 250V of UV. To provide consistency for these exposures, sample was taken from the same culture at each level of exposure. | ||
| + | Readings were taken using a microplate reader, each sample plated in triplicate. The excitation peak and emission peaks of RFP (E1010) at 584 nm and 607 nm respectively were used for the readings. Saturated culture and culture at an OD600 of approximately 0.8 with no RFP expression were used as negative controls. | ||
| + | These preliminary tests resulted in very high background readings and results that were not statistically significant as a result of this signal to noise ratio (data not shown). Repetition and protocol modification is required to continue these tests. UV seemed to have little to no effect on the induction of the pRecA promoter. Other crosslinking agents will be tested, including mitomycin C, in addtion to testing other levels of UV exposure. PocI will also be tested in the presence of the cI repressor. | ||
| - | === The | + | ==Future Directions and Applications== |
| + | *A post induction of genome degradation freeze drying system can be utilized to enable storage and transport for subsequent thawing and induction of the desired product. | ||
| + | *Quorum sensing control T7PoI can be expressed. The nuclease operon will be put under T7 control instead of TetR | ||
| + | ==References== | ||
| + | |||
| + | O'Connor,C.D., & Timmins, K.N. (1987). Highly repressible expression system for cloning genes that specify potentialy toxic proteins. Journal of Bacteriology. 169, 4457-4462. | ||
| - | + | GUZMAN, L, BELIN, D, CARSON, M. J., & BECKWITH, J (1995). Tight Regulation, Modulation, and High-Level Expression by J. of Bacteriology, 177, 4121-4130. | |
| - | + | Macdiarmid, et al. Cancer Cell 11, 431-445, May 2007 | |
Latest revision as of 04:00, 30 October 2008
| Home | The Team | The Project | Parts Submitted to the Registry | Modeling | Notebook | Sponsors |
|---|
Contents |
Project Aim and Rationale
There are many useful bioproducts that can be efficiently expressed in vivo but are either cost-prohibitive or impossible to synthesiize and purify in vitro. To address this need for a means of manufacturing such bioproducts, a great deal of work had been done to study [http://openwetware.org/wiki/Minicells minicells], genome-free sacks of cytoplasm that bud off from certain mutant bacterial strains, and from which plasmid-driven expression has previously shown to be possible. Minicells showed substantial promise for possible therapeutic applications since they could generate useful products without replicating themselves. However, for in situ delivery of their products, they must be filtered from the live bacteria from which they were released.
Transient plasmid-driven expression in minicells has been shown to last for several hours, and though this "lifetime" can be improved by use of a nutrient bath, it is limited by cytosolic resources. Once these resources are exhausted, the minicell becomes inactive and degrades.
Our project improves on the two issues described above. A bacterial cell already containing a plasmid encoding bioproduct synthesis genes, will self-destruct by degrading its own genome and transiently produce the bioproduct until cell resources have been exhausted. Having the original bacteria serve as the factory means that not only is there no live parental budding strain to purify from the culture, but also more cell resources are available to express larger quantities of bioproduct.
Genome degradation is achieved using the combined activity of a restriction endonuclease to fragment the genome and an exonuclease to hasten degradation. The gene for the protein of interest will be located on a plasmid lacking recognition sites for the endonuclease, allowing it to remain intact after genomic degradation. The plasmid genes will be expressed using the remaining cell resources until they expire. The primary application of this design would be an in situ compound production and delivery system for agricultural, industrial or therapeutic use.
Project Design
Production and Regulation of Nuclease Genes
- Endo- and exonucleases will be cloned between the lambda pR promoter and the Plac promoter. The lambda promoter will drive the production of the forward transcript for translation of the genes (which are toxic), while pLac will drive production of antisense transcripts to maximize repression of the two nucleases during uninduced conditions. Cultures will initially be grown on media containing IPTG. Expression of the unstable cI repressor (C0051) will be driven by another instance of a lacI repressable promoter. Since nuclease production is usually repressed, endogenous lacI production should be sufficient. Any antisense transcripts from the leaky Plac will not be able to compete with the strong lambda promoter during expression of the nucleases.
Construction Strategy
Biobrick Part Number Legend
- [TT] BBa_B0015
- [Pr] BBa_R0051
- [Plac] BBa_R0011
- [CI] BBa_C0051
- [T7gene6] BBa_K093001
- [PmeI] BBa_K093002
Steps in Assembly
- T7 gene 6 exonuclease will be amplified using PCR with primer-adapters for the BioBrick Prefix and Suffix.
- Pseudomonas mendocina PmeI endonuclease will be synthesized with its ORF flanked by BioBrick Prefix and Suffix.
- A nuclease construct will be constructed using crossover PCR of the nuclease gene.
- A convergent promoter system designed to flank the nuclease genes are denoted below by Construct 1 and Construct 2. Construct 2 will be added downstream of the nuclease constructs such that no sense transcript and protein will be made at this stage. Construct 1 will be added upstream in the next round of assembly. This will ensure repression of the system in the presence of CI repressor and IPTG.
- The addition of Construct 2 is a non toxic intermediate step.
- The addition of Construct 1 ensures for proper repression in the presence of IPTG and the CI repressor.
- The already assembled CI repressor under the control of Plac must be cotransformed and plated on IPTG
- The final independent construct:
- These tests will determine:
- The length of time needed for the destruction of the genome by nuclease expression
- The optimal integration site based on the ideal length of time for nuclease gene production.
- Where the REN sites are on the genome.
- The rate of degradation by the exonuclease.
Note:
- Ligation scars are not shown.
- Ribosome binding sites are present for open reading frames (T7gene6, PmeI, CI) upstream in the forward orientation.
- [forward arrow] denotes forward orientation
- [reverse arrow] denotes reverse position.
Implementation
Preliminary Assay of Genomic Degradation
- Chassis will be transformed with the preliminary construct.
- Uninduced cell growth will be assayed to determine if the repression is strong enough to allow for the uninhibited growth of uninduced cells.
- Cells will be induced for different periods of time. The induction will be terminated by washing to eliminate IPTG. Assay for sterility of culture growth test with positive and negative controls (uninduced transformants and no inoculation, respectively). Genome destruction and plasmid safeness will be checked by genomic and plasmid preps of the induced culture. [(looking for smear and nice band?)]
Integration of Nuclease Genes
- The genome degradation construct will be integrated into the genome via a proposed standard integration vector.
- The vector will contain a BioBrick cloning site and unique double digest sites flanking the BioBrick cloning site and suitable elements for selection (Sac B and antibiotic resistance). The unique double digest sites will allow the user to customize the regions of homology for a double crossover homologous recombination.
- The integration vector will be a vector with the oriR6k origin of replication that only replicates in E. coli lambda pir lysogen. For integration, the plasmid will be transformed into a strain of E. coli that is not a lambda pir lysogen by selecting for recombination using the antibiotic respective to the vector. The vector will be a suicide vector and therefore in any non lambda pir strain, a single recombination will be selected. The cells will then be removed from selective pressure to allow for the next recombination to occur. The double recombinants will be selected using sucrose, since the vector will also contain a sacB gene, which confers sucrose sensitivity. The putative double recombinants must then be screened for the respective antibiotic sensitivity.
Safe Target Plasmid
- In order to ensure expression of a desired gene post degradation of the genome, the plasmid must be free of recognition sites for the restriction endonuclease PmeI and therefore be unable to be influenced by the exonuclease. Red fluorescent protein is used as a reporter gene.
- The reporter will be put under the control of the LexA repressible promoter. The genome degradation method produces single stranded DNA due to the presence of the 5’-3’ [(check gene 6 exo activity direction to confirm)] exonuclease. When there is single stranded DNA in the cell, RecA is upregulated as part of the SOS response, and RecA cleaves lexA. Therefore when the genome degradation is induced, the expression of the gene of interest will be subsequently induced. For this reason, the final construct must be integrated into a RecA + strain (TG1 is Rec+ and lacIq, and will work completely since lacI is required for the induction of the genome degradation operon).
- The plasmid safeness will be assayed when exposed to the nucleases. From that, reporter production can be evaluated, and the induction time relative to the time of degradation can be optimized.
Preliminary Testing of Promoter Reporter Constructs
A preliminary experiment was conducted to test the expression of the pRecA promoter with LexA binding sites (K093000), the constitutive promoter pconst (J23118) and PocI, the lambda Pr promoter containing the operator for lambda cI repressor (K093010). Visual observations of liquid cultures show that expression of RFP under the control of the PocI promoter in absence of the repressor is high and similar to expression under control of the constitutive promoter. These cultures were all extremely pink, and also form pink colonies on solid media. The uninduced pRecA culture shows very low levels of expression and light pink colonies.
Cultures were inoculated in triplicate. Saturated overnight cultures were inoculated from a single colony. A second set of cultures were inoculated and grown to an OD600 of approximately 0.8. For pRecA, 3 conditions were set and conducted at both cell concentrations; uninduced culture, culture exposed to 125V of UV culture exposed to 250V of UV. To provide consistency for these exposures, sample was taken from the same culture at each level of exposure.
Readings were taken using a microplate reader, each sample plated in triplicate. The excitation peak and emission peaks of RFP (E1010) at 584 nm and 607 nm respectively were used for the readings. Saturated culture and culture at an OD600 of approximately 0.8 with no RFP expression were used as negative controls.
These preliminary tests resulted in very high background readings and results that were not statistically significant as a result of this signal to noise ratio (data not shown). Repetition and protocol modification is required to continue these tests. UV seemed to have little to no effect on the induction of the pRecA promoter. Other crosslinking agents will be tested, including mitomycin C, in addtion to testing other levels of UV exposure. PocI will also be tested in the presence of the cI repressor.
Future Directions and Applications
- A post induction of genome degradation freeze drying system can be utilized to enable storage and transport for subsequent thawing and induction of the desired product.
- Quorum sensing control T7PoI can be expressed. The nuclease operon will be put under T7 control instead of TetR
References
O'Connor,C.D., & Timmins, K.N. (1987). Highly repressible expression system for cloning genes that specify potentialy toxic proteins. Journal of Bacteriology. 169, 4457-4462.
GUZMAN, L, BELIN, D, CARSON, M. J., & BECKWITH, J (1995). Tight Regulation, Modulation, and High-Level Expression by J. of Bacteriology, 177, 4121-4130.
Macdiarmid, et al. Cancer Cell 11, 431-445, May 2007
 "
"