Team:Hawaii/Notebook/2008-08-31
From 2008.igem.org
(Difference between revisions)
(New page: {{Team:Hawaii/Header}} = Things we did today = == Wetlab work == ===Construction of p+r (cont.) and re-replacement of BB-pRL1833a MCS=== :<strong> Grace</strong> [[Image:083108REdigests.j...) |
(→Construction of p+r (cont.) and re-replacement of BB-pRL1833a MCS) |
||
| (5 intermediate revisions not shown) | |||
| Line 7: | Line 7: | ||
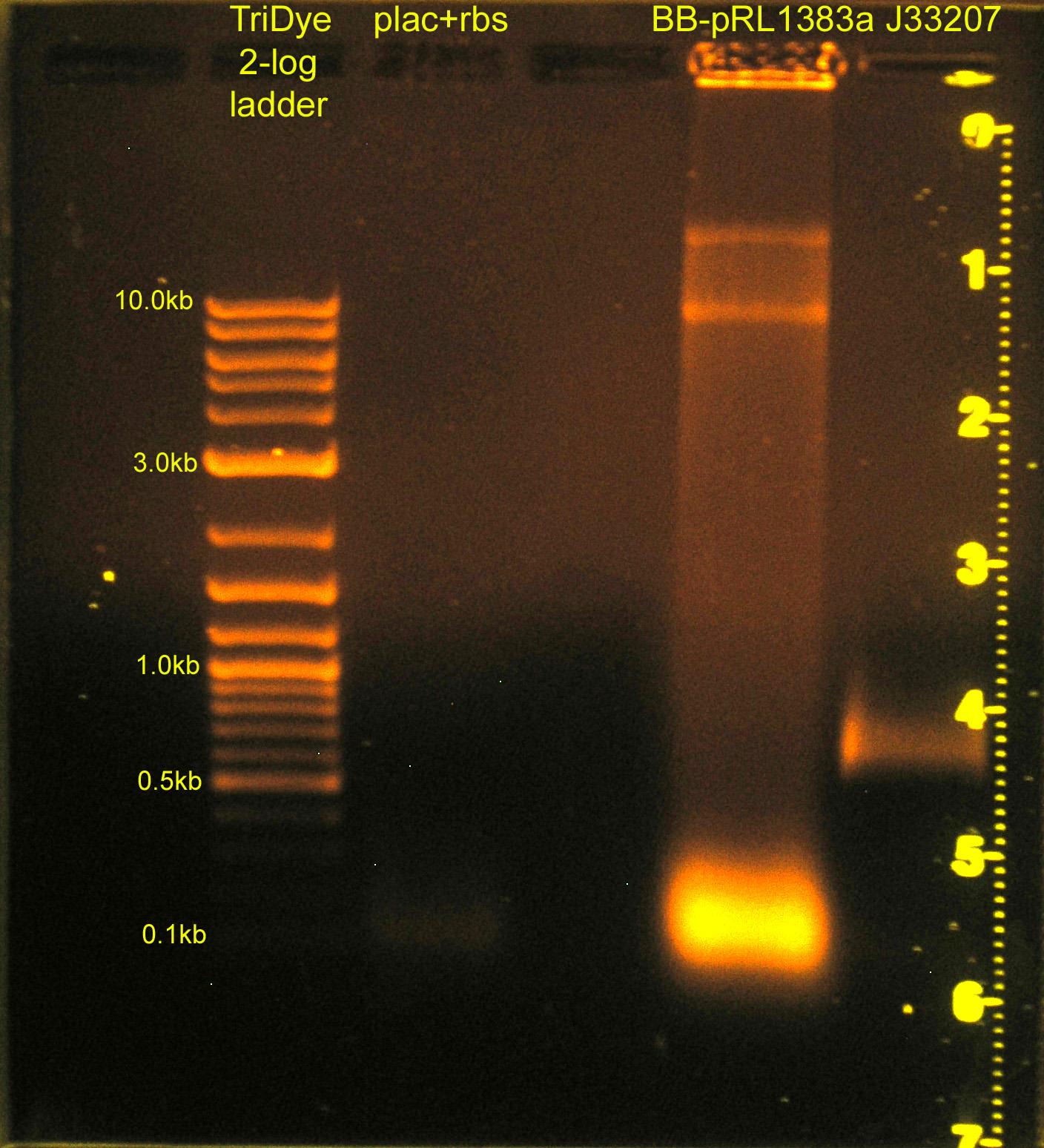
[[Image:083108REdigests.jpg|right|thumb|200px|EtBr stained 0.8% agarose gel ran at 60V for 1.5 hours.]] | [[Image:083108REdigests.jpg|right|thumb|200px|EtBr stained 0.8% agarose gel ran at 60V for 1.5 hours.]] | ||
:* Ran RE digests from last night on gel | :* Ran RE digests from last night on gel | ||
| + | ::*plac+rbs band not visible; will ligate using plac and rbs parts (instead of plac+rbs part) | ||
| + | :::*Image to right has been edited w/ Photoshop. Faint smear for plac+rbs was not visible before. | ||
:* Extracted bands from gel | :* Extracted bands from gel | ||
:* Ligated: | :* Ligated: | ||
| - | ::* plac | + | ::* plac and rbs (B0034) and C0012 vector |
::* J33207 and BB-pRL1383a | ::* J33207 and BB-pRL1383a | ||
:* Transformed into DH5α cells | :* Transformed into DH5α cells | ||
| - | ::* Plated BB-pRL1383a+J33207 on | + | ::* Plated BB-pRL1383a+J33207 on selective media with and without IPTG to verify if IPTG is necessary for blue/white screening in this strain |
| + | |||
| + | ===Inoculated TB+sp<sub>100</sub> with BB-pRL1383a=== | ||
| + | :<strong>Grace</strong> | ||
| + | |||
| + | ===Construction of Broad-Host-Range Plasmid Parts=== | ||
| + | :<strong>Margaret</strong> | ||
| + | |||
| + | [[Image:re_digest_8_31_08.jpg|right|thumb|150px|Restriction digest of rep and P1 lytic regions.]] | ||
| + | :*gel of yesterday's re-digest, lane 3 was cleaned using gel purification spin columns and stored in -20°C. Lanes 7-10 were extracted and cleaned as well. | ||
| + | :*Ligation: using T4 ligase, 10X buffer (from the same lot as the enzyme, it was aliquoted today). 2 hour incubation at room temperature. The reaction was placed in 4°C in case transformation does not yield colonies. | ||
| + | |||
| + | {|class=wikitable border=1 align=center | ||
| + | !insert | ||
| + | !vector | ||
| + | |- | ||
| + | |1 pSB1A3 cut with X,P(de-P0<sub>4</sub><sup>2-</sup>)18ng | ||
| + | |3 rep (PCR product) X,P 82ng | ||
| + | |- | ||
| + | |1pSB1A3 cut with X,P(de-P0<sub>4</sub><sup>2-</sup>)35.6ng | ||
| + | |3P1 lytic region X,P 64.4ng | ||
| + | |- | ||
| + | |3pSB1A3 cut with X,P(de-P0<sub>4</sub><sup>2-</sup>)66.2ng | ||
| + | |1 rep (PCR product) X,P 33.8ng\ | ||
| + | |} | ||
| + | |||
| + | :*Transformation: of ligation products from today into DH5-alpha cells. Everything plated on Amp100 LB plates, pSMC121 used as positive control (plated on SmSp). | ||
| + | |||
| + | :* Plasmid Prep: stored in -20°C in 30ul TE buffer | ||
| + | :*Culture: 5mL Terrific Broth + amp100 | ||
| + | ::*plac+B0030 (1:6):3, 4, 7 | ||
| + | ::*plac+B0030 :3 | ||
| + | ::*plac+B0034 (1:6):1 | ||
| + | ::*plac+B0034 (1:3):3 | ||
| + | |||
= Discussion = | = Discussion = | ||
Latest revision as of 06:39, 1 September 2008
| Projects | Events | Resources | ||
|---|---|---|---|---|
| Sponsors | Experiments | Milestones | Protocols | |
| Notebook (t) | Meetings (t) |
Things we did today
Wetlab work
Construction of p+r (cont.) and re-replacement of BB-pRL1833a MCS
- Grace
- Ran RE digests from last night on gel
- plac+rbs band not visible; will ligate using plac and rbs parts (instead of plac+rbs part)
- Image to right has been edited w/ Photoshop. Faint smear for plac+rbs was not visible before.
- Extracted bands from gel
- Ligated:
- plac and rbs (B0034) and C0012 vector
- J33207 and BB-pRL1383a
- Transformed into DH5α cells
- Plated BB-pRL1383a+J33207 on selective media with and without IPTG to verify if IPTG is necessary for blue/white screening in this strain
Inoculated TB+sp100 with BB-pRL1383a
- Grace
Construction of Broad-Host-Range Plasmid Parts
- Margaret
File:Re digest 8 31 08.jpg
Restriction digest of rep and P1 lytic regions.
- gel of yesterday's re-digest, lane 3 was cleaned using gel purification spin columns and stored in -20°C. Lanes 7-10 were extracted and cleaned as well.
- Ligation: using T4 ligase, 10X buffer (from the same lot as the enzyme, it was aliquoted today). 2 hour incubation at room temperature. The reaction was placed in 4°C in case transformation does not yield colonies.
| insert | vector |
|---|---|
| 1 pSB1A3 cut with X,P(de-P042-)18ng | 3 rep (PCR product) X,P 82ng |
| 1pSB1A3 cut with X,P(de-P042-)35.6ng | 3P1 lytic region X,P 64.4ng |
| 3pSB1A3 cut with X,P(de-P042-)66.2ng | 1 rep (PCR product) X,P 33.8ng\ |
- Transformation: of ligation products from today into DH5-alpha cells. Everything plated on Amp100 LB plates, pSMC121 used as positive control (plated on SmSp).
- Plasmid Prep: stored in -20°C in 30ul TE buffer
- Culture: 5mL Terrific Broth + amp100
- plac+B0030 (1:6):3, 4, 7
- plac+B0030 :3
- plac+B0034 (1:6):1
- plac+B0034 (1:3):3
Discussion
Quote of the Day
History is the only laboratory we have in which to test the consequences of thought. - Étienne Gilson
[http://manoa.hawaii.edu/  ][http://manoa.hawaii.edu/ovcrge/
][http://manoa.hawaii.edu/ovcrge/  ][http://www.ctahr.hawaii.edu
][http://www.ctahr.hawaii.edu  ]
]
 "
"