Team:KULeuven/Project/CellDeath
From 2008.igem.org
m |
|||
| (6 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
| + | {{:Team:KULeuven/Tools/Styling}} | ||
| + | {{:Team:KULeuven/Tools/Scripting}} | ||
{{:Team:KULeuven/Tools/Header}} | {{:Team:KULeuven/Tools/Header}} | ||
| - | + | [[Image:pictogram_celldeath.png|120px|right]] | |
==Cell Death== | ==Cell Death== | ||
| Line 12: | Line 14: | ||
[[Image:Hybrid_promotor.PNG|left]] | [[Image:Hybrid_promotor.PNG|left]] | ||
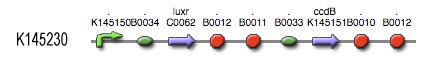
The LuxR protein [http://partsregistry.org/Part:BBa_C0062 '''BBa_C0062'''] is placed under control of our own Lux-box - c2 P22 hybrid promoter [http://partsregistry.org/Part:BBa_K145150 '''BBa_145150''']. It functions as shown schematically in the adjacent figure. The LuxR reading frame is accessible through a [http://partsregistry.org/Part:BBa_B0034 '''BBa_0034'''] RBS (relative efficiency 1.00) and is followed by [http://partsregistry.org/Part:BBa_B0014 '''BBa_B0014'''], a transcription terminator with about 60% efficiency. This means that only 40% of the engaged RNA polymerases will read through and reach the more downstream ccdB coding region [http://partsregistry.org/Part:BBa_K145151 '''BBa_K145151''']. This open reading frame can be accessed through a [http://partsregistry.org/Part:BBa_B0033 '''BBa_B0033'''] RBS (relative efficiency 0.01) and is followed by a [http://partsregistry.org/Part:BBa_B0015 '''BBa_B0015'''], highly efficient transcription terminator. | The LuxR protein [http://partsregistry.org/Part:BBa_C0062 '''BBa_C0062'''] is placed under control of our own Lux-box - c2 P22 hybrid promoter [http://partsregistry.org/Part:BBa_K145150 '''BBa_145150''']. It functions as shown schematically in the adjacent figure. The LuxR reading frame is accessible through a [http://partsregistry.org/Part:BBa_B0034 '''BBa_0034'''] RBS (relative efficiency 1.00) and is followed by [http://partsregistry.org/Part:BBa_B0014 '''BBa_B0014'''], a transcription terminator with about 60% efficiency. This means that only 40% of the engaged RNA polymerases will read through and reach the more downstream ccdB coding region [http://partsregistry.org/Part:BBa_K145151 '''BBa_K145151''']. This open reading frame can be accessed through a [http://partsregistry.org/Part:BBa_B0033 '''BBa_B0033'''] RBS (relative efficiency 0.01) and is followed by a [http://partsregistry.org/Part:BBa_B0015 '''BBa_B0015'''], highly efficient transcription terminator. | ||
| + | |||
| + | ===Action=== | ||
[[Image:LuxRHSL.png|400px|right]] | [[Image:LuxRHSL.png|400px|right]] | ||
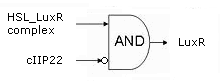
| - | + | Initially, the system is in the OFF state, with the [https://2008.igem.org/Team:KULeuven/Project/Memory Memory] stably producing c2 P22. This is the repressor for the hybrid promoter, as long as it is present, there will be no transcription of the downstream genes. Simply put: no LuxR or ccdB. | |
| - | Initially, the system is in the OFF state, with the [https://2008.igem.org/Team:KULeuven/Project/Memory | + | |
| - | When a significant and consistent [https://2008.igem.org/Team:KULeuven/Project/Input | + | When a significant and consistent [https://2008.igem.org/Team:KULeuven/Project/Input Input] signal makes the memory switch to the ON state, production of c2 P22 is halted from that point on in favor of the cI 434 repressor. The removal of c2 P22 from the system means that the hybrid promoter is no longer repressed and a small background amount of LuxR will be produced, toghether with a very tiny, sublethal amount of ccdB. The differences in amount produced are realised by the use of different Ribosome Binding Sites ([http://partsregistry.org/Part:BBa_B0034 '''BBa_B0034'''] in front of ''luxR'' vs the 100x less efficient [http://partsregistry.org/Part:BBa_B0033 '''BBa_B0033'''] preceding ''ccdB'') and the presence of a transcription terminator in front of the ''ccdB'' gene. |
| - | As long as this input signal remains, the [https://2008.igem.org/Team:KULeuven/Project/Inverter | + | As long as this input signal remains, the [https://2008.igem.org/Team:KULeuven/Project/Inverter InverTimer] produces no 3OC6HSL and transcription from the hybrid promoter will remain on the background level. |
| - | If the input signal fades away however, so does Lactonase ([https://2008.igem.org/Team:KULeuven/Project/Reset | + | If the input signal fades away however, so does Lactonase ([https://2008.igem.org/Team:KULeuven/Project/Reset Reset]) while the [https://2008.igem.org/Team:KULeuven/Project/Inverter InverTimer] starts making LuxI and thus 3OC6HSL. |
If high enough levels of HSL are reached, it can associate with LuxR, enabling the complex to dimerise, bind the Lux-box in the hybrid promoter and upregulate transription of the downstream genes. In this way, more LuxR is produced in an auto-activating manner and the system evolves to the fully activated state. Then, and only then will a significant (read: lethal) amount of ccdB be produced and the cell dies ([http://www.ncbi.nlm.nih.gov/pubmed/10196173?dopt=Abstract reference]). | If high enough levels of HSL are reached, it can associate with LuxR, enabling the complex to dimerise, bind the Lux-box in the hybrid promoter and upregulate transription of the downstream genes. In this way, more LuxR is produced in an auto-activating manner and the system evolves to the fully activated state. Then, and only then will a significant (read: lethal) amount of ccdB be produced and the cell dies ([http://www.ncbi.nlm.nih.gov/pubmed/10196173?dopt=Abstract reference]). | ||
Latest revision as of 12:28, 5 October 2008
Contents |
Cell Death
BioBricks
Components
The LuxR protein [http://partsregistry.org/Part:BBa_C0062 BBa_C0062] is placed under control of our own Lux-box - c2 P22 hybrid promoter [http://partsregistry.org/Part:BBa_K145150 BBa_145150]. It functions as shown schematically in the adjacent figure. The LuxR reading frame is accessible through a [http://partsregistry.org/Part:BBa_B0034 BBa_0034] RBS (relative efficiency 1.00) and is followed by [http://partsregistry.org/Part:BBa_B0014 BBa_B0014], a transcription terminator with about 60% efficiency. This means that only 40% of the engaged RNA polymerases will read through and reach the more downstream ccdB coding region [http://partsregistry.org/Part:BBa_K145151 BBa_K145151]. This open reading frame can be accessed through a [http://partsregistry.org/Part:BBa_B0033 BBa_B0033] RBS (relative efficiency 0.01) and is followed by a [http://partsregistry.org/Part:BBa_B0015 BBa_B0015], highly efficient transcription terminator.
Action
Initially, the system is in the OFF state, with the Memory stably producing c2 P22. This is the repressor for the hybrid promoter, as long as it is present, there will be no transcription of the downstream genes. Simply put: no LuxR or ccdB.
When a significant and consistent Input signal makes the memory switch to the ON state, production of c2 P22 is halted from that point on in favor of the cI 434 repressor. The removal of c2 P22 from the system means that the hybrid promoter is no longer repressed and a small background amount of LuxR will be produced, toghether with a very tiny, sublethal amount of ccdB. The differences in amount produced are realised by the use of different Ribosome Binding Sites ([http://partsregistry.org/Part:BBa_B0034 BBa_B0034] in front of luxR vs the 100x less efficient [http://partsregistry.org/Part:BBa_B0033 BBa_B0033] preceding ccdB) and the presence of a transcription terminator in front of the ccdB gene.
As long as this input signal remains, the InverTimer produces no 3OC6HSL and transcription from the hybrid promoter will remain on the background level.
If the input signal fades away however, so does Lactonase (Reset) while the InverTimer starts making LuxI and thus 3OC6HSL. If high enough levels of HSL are reached, it can associate with LuxR, enabling the complex to dimerise, bind the Lux-box in the hybrid promoter and upregulate transription of the downstream genes. In this way, more LuxR is produced in an auto-activating manner and the system evolves to the fully activated state. Then, and only then will a significant (read: lethal) amount of ccdB be produced and the cell dies ([http://www.ncbi.nlm.nih.gov/pubmed/10196173?dopt=Abstract reference]).
The latter scenario occurs of course only if the cell isn't saved by a new sigificant input signal which resets the timer and brings the LuxR and ccdB transcription back to their background levels.
 "
"


 Input
Input Output
Output Filter
Filter InverTimer
InverTimer Reset
Reset Cell Death
Cell Death Memory
Memory