Team:UC Berkeley/GatewayGenomic
From 2008.igem.org
JCAnderson (Talk | contribs) (→Discussion) |
(→General Procedure) |
||
| (33 intermediate revisions not shown) | |||
| Line 4: | Line 4: | ||
==Introduction== | ==Introduction== | ||
| - | The ''in vivo'' plasmid based gateway scheme was successful and | + | The ''in vivo'' plasmid based gateway scheme was successful and produced the desired product, but it also yielded a considerable amount of background. Closer analysis of the side products revealed that the background resulted from ''ccdB'' mutations that arose when the gene replicated and recombined ''in vivo''. In an effort to circumvent background resulting from mutations in a negative selection gene, we decided to use positive selection. This way, we would be able to select for the desired products rather than selecting against unwanted side products and unreacted starting materials. |
| - | In order to implement positive selection, we used | + | In order to implement positive selection, we used conditional origins of replication on our plasmids. These origins of replication will only be functional and allow plasmid replication if a specific protein is expressed in the cell. In our design, the assembly vector with an ''oriR6K'' origin (requires the protein Pi) is integrated into the genome. The gene of interest alongside an ''oriV'' origin (requires the protein TrfA) can then recombine with the genome. In this manner, the gene of interest can be moved to an assembly vector by inserting the entry vector into the genome. We could allow selective replication of the plasmids in our scheme by performing the reaction in a cell expressing TrfA and then transforming the resulting mixture of plasmids into a cell expressing ‘’pir’’. |
We also incorporated our lysis device into our new entry vector in order to simplify the experimental protocol for this scheme by eliminating mini-preps. | We also incorporated our lysis device into our new entry vector in order to simplify the experimental protocol for this scheme by eliminating mini-preps. | ||
| Line 13: | Line 13: | ||
===Plasmids=== | ===Plasmids=== | ||
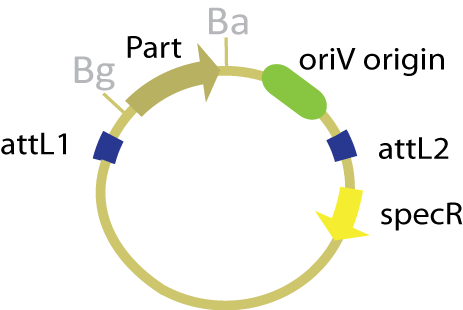
| + | The entry plasmid in this scheme contains both the part of interest and an ''oriV'' origin of replication within the attL recombination sites, thereby allowing this origin to be transferred to the desired product. Since the entry plasmid has no constitutive origin of replication, it can only replicate in the presence of the protein TrfA. With the exception of the change in type and placement of the origin of replication, the entry plasmid maintains the same features as the traditional entry plasmid, pBca1256. | ||
| + | [[Image:OriV entry vector.jpg|250 px]] | ||
| + | |||
| + | In variations on this scheme where Xis and Int are not integrated into the genome, they are placed prior to the'' attL1'' site under the control of a temperature sensitive promoter. In an effort to streamline this method, we also considered a variation where the lysis device was placed before Xis and Int in the entry plasmid. | ||
===Strains=== | ===Strains=== | ||
| - | This scheme utilizes strains that include the assembly vector in the genome. The traditional double-antibiotic assembly vector has been modified to include an | + | This scheme utilizes strains that include the assembly vector in the genome. The traditional double-antibiotic assembly vector has been modified to include an ''oriR6K'' origin of replication (which can be induced by the Pi protein) in place of a constitutively active replication origin. Since the integrated vector includes the ''attR2'' sites that usually flank the vector region of the assembly plasmid, the ''attL1'' sites in the entry plasmid can recombine with the genome to produce the desired product. |
| + | |||
| + | In addition to the integrated assembly vector, the strains also include TrfA in the genome to allow the replication of the entry vector containing an ''oriV'' origin. Additionally, one variation on this scheme has ''xis'' and ''int'' placed in the the genome to catalyze the recombination reaction. | ||
| + | |||
| + | ==General Procedure== | ||
| + | In this scheme, the ''oriV'' version of the entry plasmid can simply be transformed into the integrated strain containing the desired assembly vector and TrfA. In order to catalyze the reaction, Xis and Int must be present either on the plasmid or in the genome. | ||
| + | |||
| + | Once the entry plasmid has been transformed and the reaction has been catalyzed, the desired product will be able to replicate in the cytoplasm of the cell using the ''oriV'' origin that was transferred with the part from the entry vector to the assembly vector. The product can be released from the cell by inducing our self-lysis device (which can be installed in the entry vector) or by doing a mini-prep. | ||
| + | |||
| + | Once the plasmids in the cell's cytoplasm have been obtained, they need to be transformed into cells that express the Pi protein. This will allow for the selective replication of the desired product, which contains an ''oriR6K'' origin obtained from the assembly vector that was originally in the genome. | ||
| + | |||
| + | The potential source of background in this scheme is the unreacted entry plasmid. However, this plasmid does not have an ''oriR6K'' origin (it only contains an ''oriV'' origin), so it is unable to replicate in cells that express Pi instead of TrfA. Differential antibiotic markers would assist in screening against the bleed-through of the starting plasmid, but only positive selection can eliminate the need to screen for contransformants (which contain both the starting plasmid in addition to the desired product). | ||
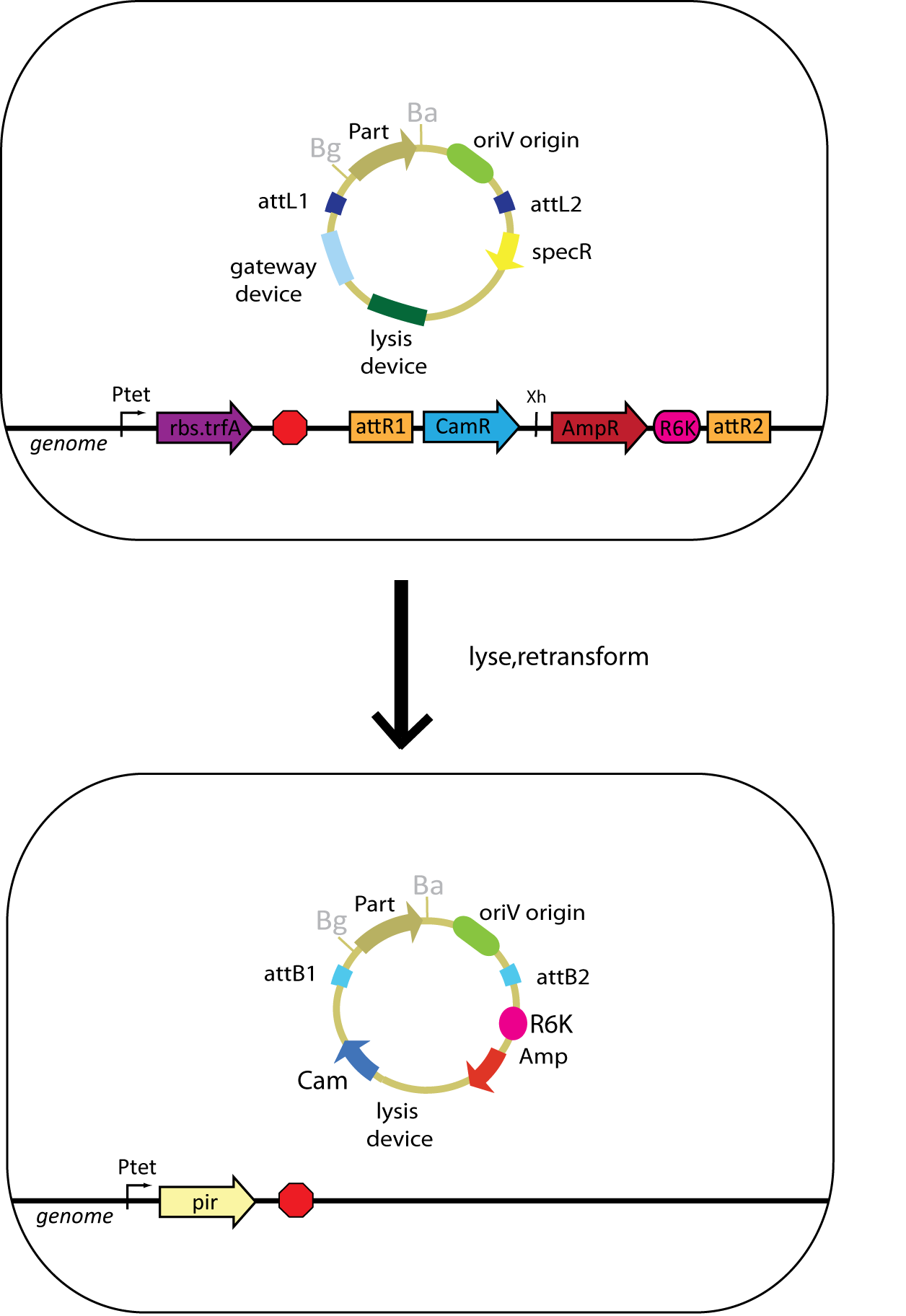
| - | + | [[Image:Genomic Gateway Overview6.jpg|500 px]] | |
| - | == | + | ==Conclusion== |
| + | With the incorporation of the lysis device, the genomic Gateway scheme promises to be an improvement to the plasmid based Gateway scheme because it eliminates unwanted background by using positive selection. However, the protocol for this scheme still requires that the plasmid mixture is transformed into another cell strain to select for the desired product. We sought to further streamline this scheme by utilizing phagemids, which can lyse the cell and infect other cells without the need for either mini-preps or transformation. | ||
| - | |||
<html> | <html> | ||
Latest revision as of 04:14, 30 October 2008
Introduction
The in vivo plasmid based gateway scheme was successful and produced the desired product, but it also yielded a considerable amount of background. Closer analysis of the side products revealed that the background resulted from ccdB mutations that arose when the gene replicated and recombined in vivo. In an effort to circumvent background resulting from mutations in a negative selection gene, we decided to use positive selection. This way, we would be able to select for the desired products rather than selecting against unwanted side products and unreacted starting materials.
In order to implement positive selection, we used conditional origins of replication on our plasmids. These origins of replication will only be functional and allow plasmid replication if a specific protein is expressed in the cell. In our design, the assembly vector with an oriR6K origin (requires the protein Pi) is integrated into the genome. The gene of interest alongside an oriV origin (requires the protein TrfA) can then recombine with the genome. In this manner, the gene of interest can be moved to an assembly vector by inserting the entry vector into the genome. We could allow selective replication of the plasmids in our scheme by performing the reaction in a cell expressing TrfA and then transforming the resulting mixture of plasmids into a cell expressing ‘’pir’’.
We also incorporated our lysis device into our new entry vector in order to simplify the experimental protocol for this scheme by eliminating mini-preps.
Details about Plasmids and Strains
Plasmids
The entry plasmid in this scheme contains both the part of interest and an oriV origin of replication within the attL recombination sites, thereby allowing this origin to be transferred to the desired product. Since the entry plasmid has no constitutive origin of replication, it can only replicate in the presence of the protein TrfA. With the exception of the change in type and placement of the origin of replication, the entry plasmid maintains the same features as the traditional entry plasmid, pBca1256.
In variations on this scheme where Xis and Int are not integrated into the genome, they are placed prior to the attL1 site under the control of a temperature sensitive promoter. In an effort to streamline this method, we also considered a variation where the lysis device was placed before Xis and Int in the entry plasmid.
Strains
This scheme utilizes strains that include the assembly vector in the genome. The traditional double-antibiotic assembly vector has been modified to include an oriR6K origin of replication (which can be induced by the Pi protein) in place of a constitutively active replication origin. Since the integrated vector includes the attR2 sites that usually flank the vector region of the assembly plasmid, the attL1 sites in the entry plasmid can recombine with the genome to produce the desired product.
In addition to the integrated assembly vector, the strains also include TrfA in the genome to allow the replication of the entry vector containing an oriV origin. Additionally, one variation on this scheme has xis and int placed in the the genome to catalyze the recombination reaction.
General Procedure
In this scheme, the oriV version of the entry plasmid can simply be transformed into the integrated strain containing the desired assembly vector and TrfA. In order to catalyze the reaction, Xis and Int must be present either on the plasmid or in the genome.
Once the entry plasmid has been transformed and the reaction has been catalyzed, the desired product will be able to replicate in the cytoplasm of the cell using the oriV origin that was transferred with the part from the entry vector to the assembly vector. The product can be released from the cell by inducing our self-lysis device (which can be installed in the entry vector) or by doing a mini-prep.
Once the plasmids in the cell's cytoplasm have been obtained, they need to be transformed into cells that express the Pi protein. This will allow for the selective replication of the desired product, which contains an oriR6K origin obtained from the assembly vector that was originally in the genome.
The potential source of background in this scheme is the unreacted entry plasmid. However, this plasmid does not have an oriR6K origin (it only contains an oriV origin), so it is unable to replicate in cells that express Pi instead of TrfA. Differential antibiotic markers would assist in screening against the bleed-through of the starting plasmid, but only positive selection can eliminate the need to screen for contransformants (which contain both the starting plasmid in addition to the desired product).
Conclusion
With the incorporation of the lysis device, the genomic Gateway scheme promises to be an improvement to the plasmid based Gateway scheme because it eliminates unwanted background by using positive selection. However, the protocol for this scheme still requires that the plasmid mixture is transformed into another cell strain to select for the desired product. We sought to further streamline this scheme by utilizing phagemids, which can lyse the cell and infect other cells without the need for either mini-preps or transformation.
 "
"