Team:Freiburg Cloning Strategy
From 2008.igem.org
| (One intermediate revision not shown) | |||
| Line 8: | Line 8: | ||
<h2>Introduction</h2> | <h2>Introduction</h2> | ||
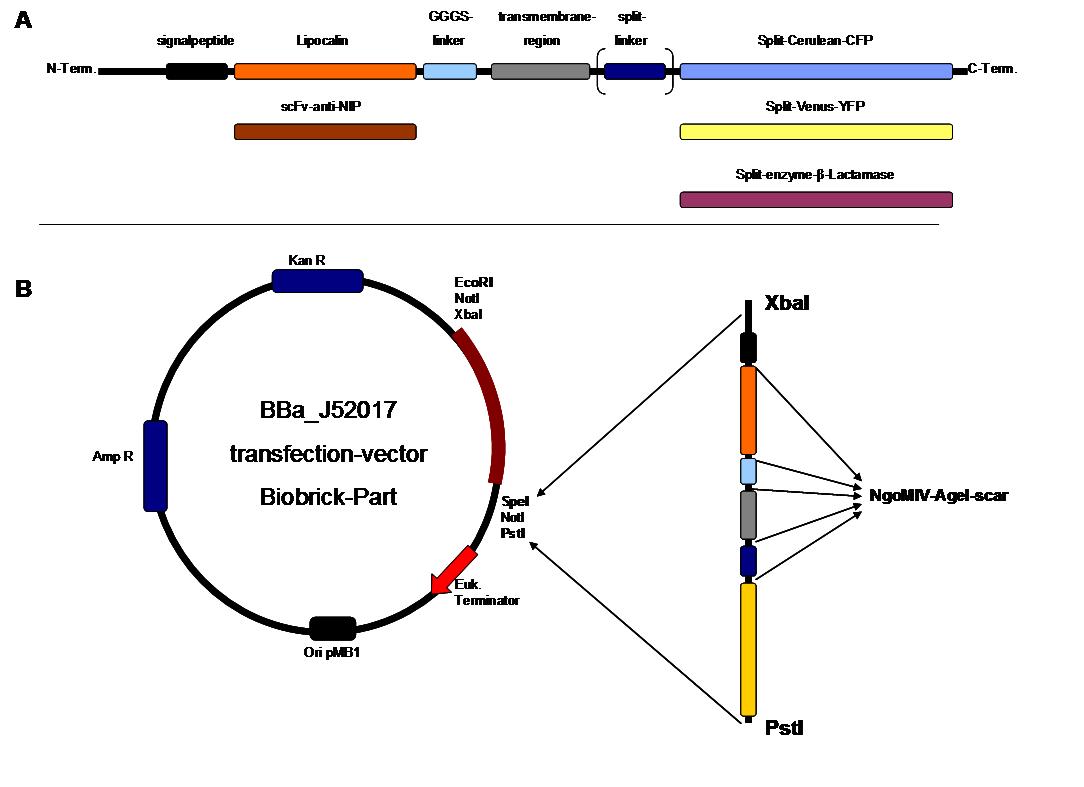
| - | To form different types of synthetic receptor constructs a | + | To form different types of synthetic receptor constructs a modular building set was used. We designed the genes of two parts coding for extracellular binding proteins (anti NIP scFv, anti-Flu Lipocalin) and four different components of an intracellular signal transduction reporter protein (Cerulean CFP, Venus YFP, β-Lactamase, and Luciferase). These reporters were designed as split-proteins to achieve activaton only when two receptors come together and form a cluster. With this strategy, a signal is only given by two binding-molecules that are next to each other as it is realized in the DNA-origami structure or the NIP and Fluorescein-coupled BSA. <br> |
| - | The whole receptor construct consists out of (i) a signal peptide, (ii) an extracellular receptor domain, (iii) a GGGS-linker followed by (iv) a transmembrane region and (v) an intracellular split-reporter-protein. The signal peptide sequence | + | The whole receptor construct consists out of (i) a signal peptide, (ii) an extracellular receptor domain, (iii) a GGGS-linker followed by (iv) a transmembrane region and (v) an intracellular split-reporter-protein. The signal peptide sequence equal to that of human EGF-receptor erbb1 ensures the transport of the protein construct to the cytoplasma membrane. A GGGS-linker is necessary to create a distance between membrane and the recognition-site to tower cell surface structures as glycoproteins and glycolipids. The sequence of the transmembrane region is taken from the EGF-receptor erbb1 as well. The C-terminal split-parts of Venus-YFP and Cerulean CFP were not directly fused to the transmembrane region. To get a little bit more flexibility in binding to the N-terminal part, a split fluorophor-linker was assembled in between.<br><br> |
| - | Almost all parts were ordered by gene synthesis in a pMA-vector system that is adapted for cloning BioBrick parts. The constructs were designed according to BioBrick 3.0 standard with the modification for fusion-proteins proposed by the iGEM Freiburg Team 2007 ([ | + | Almost all parts were ordered by gene synthesis in a pMA-vector system that is adapted for cloning BioBrick parts. The constructs were designed according to BioBrick 3.0 standard with the modification for fusion-proteins proposed by the iGEM Freiburg Team 2007 ([https://2007.igem.org/Freiburg07/report_fusion_parts FreiGEM07_report_fusion_part]). Keeping the standard iGEM prefix and suffix, the restriction sites NgoMIV and AgeI were added to create functional fusions without stop-codons. <br> |
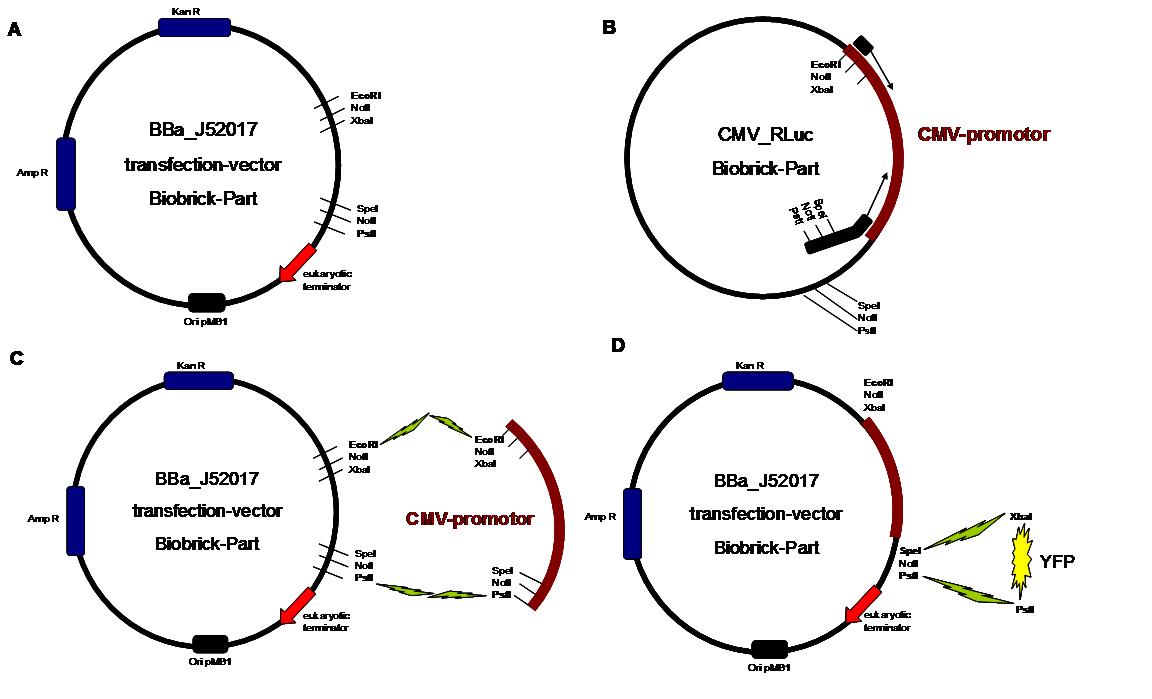
All possible variations of the receptor constructs were fused together in the pMA-vector and afterwards inserted into a transfection vector. The transfection vector is a BioBrick 2006 part from the Ljubljana, Slovenia team ([http://partsregistry.org/Part:BBa_J52017 BBa_J52017]) and includes a kanamycin and ampicillin resistance cassette. There is a pMB1 Ori and the multiple cloning site consist of the BioBrick Prefix and Suffix restriction enzymes (Prefix: EcoRI, NotI, XbaI/ Suffix: SpeI, NotI, PstI) followed by an eukaryotic terminator sequence. To achieve a strong expression, a constitutive promoter construct was needed. Because of this, the Cytomegalovirus [CMV] promotor was amplified by PCR using the BioBrick part [http://partsregistry.org/Part:BBa_J52038 BBa_J52038 ]from the 2006 Ljubljana, Slovenia team as well and primers mentioned below. The forward primer bound in the iGEM-prefix, while the reverse primer bound at the end of the CMV-coding region. The reverse primer also contained an overhang to get the iGEM-suffix directly to the end of the promoter sequence which later allowed the cloning into the transfection vector. The CMV promoter was cloned into the vector by using the EcoRI and PstI restriction sites. To test the expression activity of the vector with the CMV promoter, the gene of the yellow fluorescent protein [YFP] was cloned downstream of the promoter region by using XbaI, PstI restriction enzymes for the YFP insert and SpeI, PstI for the vector to open the Biobrick suffix. The transfection of the resulting plasmid into 293T cells shows full functionality.<br> | All possible variations of the receptor constructs were fused together in the pMA-vector and afterwards inserted into a transfection vector. The transfection vector is a BioBrick 2006 part from the Ljubljana, Slovenia team ([http://partsregistry.org/Part:BBa_J52017 BBa_J52017]) and includes a kanamycin and ampicillin resistance cassette. There is a pMB1 Ori and the multiple cloning site consist of the BioBrick Prefix and Suffix restriction enzymes (Prefix: EcoRI, NotI, XbaI/ Suffix: SpeI, NotI, PstI) followed by an eukaryotic terminator sequence. To achieve a strong expression, a constitutive promoter construct was needed. Because of this, the Cytomegalovirus [CMV] promotor was amplified by PCR using the BioBrick part [http://partsregistry.org/Part:BBa_J52038 BBa_J52038 ]from the 2006 Ljubljana, Slovenia team as well and primers mentioned below. The forward primer bound in the iGEM-prefix, while the reverse primer bound at the end of the CMV-coding region. The reverse primer also contained an overhang to get the iGEM-suffix directly to the end of the promoter sequence which later allowed the cloning into the transfection vector. The CMV promoter was cloned into the vector by using the EcoRI and PstI restriction sites. To test the expression activity of the vector with the CMV promoter, the gene of the yellow fluorescent protein [YFP] was cloned downstream of the promoter region by using XbaI, PstI restriction enzymes for the YFP insert and SpeI, PstI for the vector to open the Biobrick suffix. The transfection of the resulting plasmid into 293T cells shows full functionality.<br> | ||
An overview about the cloning is given in table 1<br> | An overview about the cloning is given in table 1<br> | ||
| Line 1,041: | Line 1,041: | ||
The present BioBrick prefix and suffix rules are not compatible with modular protein design. Thus as in 2007, we proposed an extension of the present standard for fusion proteins in which two restriction sites are added in frame adjacent to the coding sequence. The basic parts and as well all composite parts follow this strategy. | The present BioBrick prefix and suffix rules are not compatible with modular protein design. Thus as in 2007, we proposed an extension of the present standard for fusion proteins in which two restriction sites are added in frame adjacent to the coding sequence. The basic parts and as well all composite parts follow this strategy. | ||
| - | To get further information see [ | + | To get further information see [https://2007.igem.org/Freiburg07/report_fusion_parts FreiGEM07_report_fusion_part]<br> |
Latest revision as of 09:01, 16 November 2008
|
_cloning strategy
IntroductionTo form different types of synthetic receptor constructs a modular building set was used. We designed the genes of two parts coding for extracellular binding proteins (anti NIP scFv, anti-Flu Lipocalin) and four different components of an intracellular signal transduction reporter protein (Cerulean CFP, Venus YFP, β-Lactamase, and Luciferase). These reporters were designed as split-proteins to achieve activaton only when two receptors come together and form a cluster. With this strategy, a signal is only given by two binding-molecules that are next to each other as it is realized in the DNA-origami structure or the NIP and Fluorescein-coupled BSA. Almost all parts were ordered by gene synthesis in a pMA-vector system that is adapted for cloning BioBrick parts. The constructs were designed according to BioBrick 3.0 standard with the modification for fusion-proteins proposed by the iGEM Freiburg Team 2007 (FreiGEM07_report_fusion_part). Keeping the standard iGEM prefix and suffix, the restriction sites NgoMIV and AgeI were added to create functional fusions without stop-codons.
Table1_cloning strategy overview about the cloning steps to create the different types of synthetic receptors. To get further information about the composite parts see https://2008.igem.org/Team:Freiburg/Parts
Step 1 Vector digestion: EcoRI + PstI Insert
digestion: EcoRI + PstI BBa-J52017 _CMV-promotor Step
2 Vector digestion: AgeI+SpeI Insert
digestion: NgoMIV+SpeI pMA-BBFR
_ SPLIT-Linker C-YFP C-CFP Step
3 Vector digestion: AgeI+SpeI Insert
digestion: NgoMIV+SpeI pMA-BBFR
_egfR-Tm _ N-β-Lactamase _ C-β-Lactamase _ SPLIT-Linker_ C-YFP _ N-YFP _ SPLIT-Linker_ C-CFP _ N-CFP _ BB058 (Luciferase) _ BB057 (Luciferase) Step
4 Vector digestion: AgeI+SpeI Insert
digestion: NgoMIV+SpeI pMA-BBFR
_SP _scFv-anti-NIP _ Lipocalin Step
5 Vector digestion: AgeI+SpeI Insert
digestion: NgoMIV+SpeI pMA-BBFR
_SP_ scFv-anti-NIP and pMA-BBFR-+SP_ Lipocalin _GGGS-linker (produced by Klenow fill in) Step
6 Vector digestion: AgeI+SpeI Insert
digestion: NgoMIV+SpeI pMA-BBFR
_SP_ scFv-anti-NIP _ GGGS-Li and pMA-BBFR
_ SP_ Lipocalin _ GGGS-Li _
egfR-Tm _ N-β-Lactamase _
egfR-Tm _ C-β-Lactamase _
egfR-Tm _ SPLIT-Linker_ C-YFP _
egfR-Tm _ N-YFP _
egfR-Tm _ SPLIT-Linker_ C-CFP _
egfR-Tm _ N-CFP _
egfR-Tm _ BB058 (Luciferase) _
egfR-Tm _ BB057 (Luciferase) Step
7 Vector digestion:
SpeI + PstI Insert
digestion: XbaI + PstI BBa-J52017_CMV _SP_ scFv-anti-NIP_GGGS-Li_egfR-Tm_N-β-Lactamase _ SP_ scFv-anti-NI _GGGS-Li_ egfR-Tm_C-β-Lactamase _ SP_ scFv-anti-NIP_GGGS-Li_
egfR-Tm_SPLIT-Linker_C-YFP _ SP_ scFv-anti-NIP_GGGS-Li_ egfR-Tm_N-YFP _ SP_ scFv-anti-NIP_GGGS-Li_
egfR-Tm_SPLIT-Linker_C-CFP _ SP_ scFv-anti-NIP_GGGS-Li_ egfR-Tm_N-CFP _ SP_ scFv-anti-NIP_GGGS-Li _ egfR-Tm_BB058 (Luciferase) _ SP_ scFv-anti-NIP_GGGS-Li _ egfR-Tm_BB057 (Luciferase) _ SP_ Lipocalin _GGGS-Li_ egfR-Tm_N-β-Lactamase _ SP_ Lipocalin _GGGS-Li_ egfR-Tm_C-β-Lactamase _ SP_ Lipocalin _GGGS-Li_
egfR-Tm_SPLIT-Linker_ C-YFP _ SP_ Lipocalin _GGGS-Li_
egfR-Tm_N-YFP _ SP_ Lipocalin _GGGS-Li_
egfR-Tm_SPLIT-Linker_ C-CFP _ SP_ Lipocalin _GGGS-Li_
egfR-Tm_N-CFP _ SP_ Lipocalin _GGGS-Li__ egfR-Tm _ BB058 (Luciferase) _ SP_ Lipocalin _GGGS-Li__ egfR-Tm _ BB057 (Luciferase) - about 2µg Plasmid-Prep in 20µl - 10µl volume of vector and insert DNA (about 50ng vector-DNA) - about 0.5 µg Plasmid-DNA in 5µl - Competent cells (100µl) werde defrosted on ice - 25pmol forward primer All composite parts were succesfully cloned and controlled by sequencing and transfection as well (Freiburg Transfection and Synthetic Receptor)
The present BioBrick prefix and suffix rules are not compatible with modular protein design. Thus as in 2007, we proposed an extension of the present standard for fusion proteins in which two restriction sites are added in frame adjacent to the coding sequence. The basic parts and as well all composite parts follow this strategy.
To get further information see FreiGEM07_report_fusion_part The cloning parts scFv-anti-NIP, lipocalin-FluA, Split-Venus-N-YFP, Split-Venus-C-YFP, Cerulean-N-CFP and Cerulean-C-CFP were obtained by genesynthesis and ordered by the GENEART company. The β-Lactamase fragments were adopted from genesynthesis GENEART order in 2007. The erbb1-transmembrane, Split-Fluorophor-Linker and the erbb1-signalpeptide were synthesized by ATG:biosynthetics. The CMV promoter cloning part was produced by PCR with the template J52038 using the CMV_RLuc_Forward-primer and CMV_RLuc_Reverse-primer. To synthesize the GGGS-linker a Klenow-fill-in-reaction was performed by using Klenow-fragment without exonuclease activity and GGGS-forward and reverse primer. |
 "
"